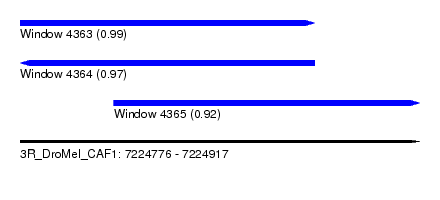
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,224,776 – 7,224,917 |
| Length | 141 |
| Max. P | 0.992772 |

| Location | 7,224,776 – 7,224,880 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.13 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
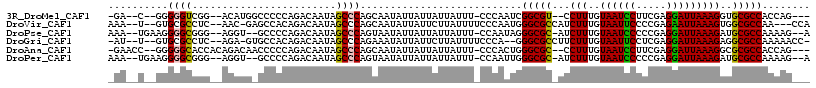
>3R_DroMel_CAF1 7224776 104 + 27905053 -GA--C--GGGGGUCGG--ACAUGGCCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU-CCCAAUCGGCGU--CCUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAG--- -..--.--((((((((.--...))))))))...............................-.......(((((--((((..((((((.....)))))))))).))))).....--- ( -34.80) >DroVir_CAF1 68802 107 + 1 AAA--U--GUGCGCCUC--AAC-GAGCCACAGACAAUAGCCCAGCAAUAUUAUUCUUAUUUUCCCAAUGGGCGCCAUCUUUGUAAUUCCCCGAGAAUUAAAGUGGCGCCAA---CCA ...--(--(((.((...--...-..))))))......................................(((((((.(((..((((((.....))))))))))))))))..---... ( -24.50) >DroPse_CAF1 91793 107 + 1 AAA--UGAAGGGGCGGG--AGGU--GCCCCAGACAAUAGCCCAGUAAUAUUAUUAUUAUUU-CCAAUAGGGCGC-AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG--A ...--....((((((..--...)--)))))............(((((((....))))))).-.......(((((-(((((..((((((.....)))))))))))))))).....--. ( -36.70) >DroGri_CAF1 92190 106 + 1 -AU--U--GUGCGCCUC--AGA-GUGCCACAGACAAUAGCCCAGAAAUAUUAUUCUUAUUUUCCCA--GGGCGCCUUCUUUGUAAUUCCUCGAGGAUUAAAGAGGCGCCAAAAACC- -.(--(--(.(((((((--((.-(((((...((.((((....((((......)))))))).))...--.)))))))((((((........)))))).....)))))))))).....- ( -31.60) >DroAna_CAF1 55870 108 + 1 -GAACC--GGGGGCACCACAGACAACCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU-CCCACUGGGCGC--CCUUUGUAAUCCUUCGAGGAUUAAAGGCGCGCCACCAG--- -.....--(((((............)))))........((((((.................-....))))))((--((((..((((((.....)))))))))).))........--- ( -32.00) >DroPer_CAF1 94872 107 + 1 AAA--UGAAGGGGCGGG--AGGU--GCCCCAGACAAUAGCCCAGUAAUAUUAUUAUUAUUU-CCAAUUGGGCGC-AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG--A ...--....((((((..--...)--)))))...((((.(...(((((((....))))))).-.).))))(((((-(((((..((((((.....)))))))))))))))).....--. ( -38.60) >consensus _AA__U__GGGGGCCGC__AGAUG_GCCCCAGACAAUAGCCCAGCAAUAUUAUUAUUAUUU_CCCAAUGGGCGC_AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG___ ..........((((........................))))...........................(((((...(((..((((((.....)))))))))..)))))........ (-18.43 = -18.13 + -0.30)
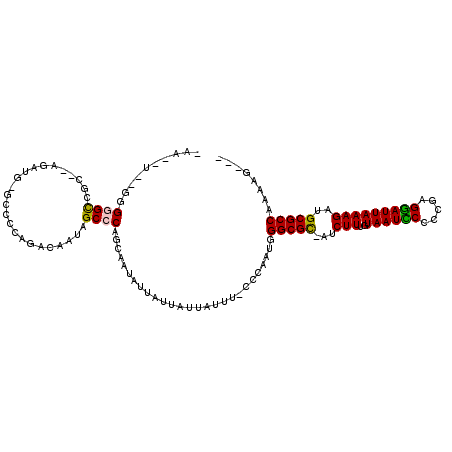

| Location | 7,224,776 – 7,224,880 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -35.31 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.26 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7224776 104 - 27905053 ---CUGGUGGCGCACCUUUAAUCCUCGAAGGAUUACAAAGG--ACGCCGAUUGGG-AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGCCAUGU--CCGACCCCC--G--UC- ---(..((((((..((((((((((.....))))))..))))--.)))).))..).-...................((((....))))(((((.(....--..).)))))--.--..- ( -32.60) >DroVir_CAF1 68802 107 - 1 UGG---UUGGCGCCACUUUAAUUCUCGGGGAAUUACAAAGAUGGCGCCCAUUGGGAAAAUAAGAAUAAUAUUGCUGGGCUAUUGUCUGUGGCUC-GUU--GAGGCGCAC--A--UUU .((---(.((((((((((((((((.....))))))..))).))))))).)))...................(((.(((((((.....)))))))-((.--...))))).--.--... ( -30.00) >DroPse_CAF1 91793 107 - 1 U--CUUUUGGCGCAUCUUUAAUCCUCGGGGGAUUACAAAGAU-GCGCCCUAUUGG-AAAUAAUAAUAAUAUUACUGGGCUAUUGUCUGGGGC--ACCU--CCCGCCCCUUCA--UUU .--.....((((((((((((((((.....))))))..)))))-)))))....(((-(..................((((....))))(((((--....--...)))))))))--... ( -37.30) >DroGri_CAF1 92190 106 - 1 -GGUUUUUGGCGCCUCUUUAAUCCUCGAGGAAUUACAAAGAAGGCGCCC--UGGGAAAAUAAGAAUAAUAUUUCUGGGCUAUUGUCUGUGGCAC-UCU--GAGGCGCAC--A--AU- -.....((((((((((.....(((....)))....((.(((....((((--.(((((.((.......)).))))))))).....))).))....-...--))))))).)--)--).- ( -30.40) >DroAna_CAF1 55870 108 - 1 ---CUGGUGGCGCGCCUUUAAUCCUCGAAGGAUUACAAAGG--GCGCCCAGUGGG-AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGGUUGUCUGUGGUGCCCCC--GGUUC- ---..(((((((((((((((((((.....))))))...)))--))))((((..(.-..............)..))))))))))..((((((((..........))))))--))...- ( -44.26) >DroPer_CAF1 94872 107 - 1 U--CUUUUGGCGCAUCUUUAAUCCUCGGGGGAUUACAAAGAU-GCGCCCAAUUGG-AAAUAAUAAUAAUAUUACUGGGCUAUUGUCUGGGGC--ACCU--CCCGCCCCUUCA--UUU .--.....((((((((((((((((.....))))))..)))))-)))))....(((-(..................((((....))))(((((--....--...)))))))))--... ( -37.30) >consensus ___CUUUUGGCGCAUCUUUAAUCCUCGAGGGAUUACAAAGAU_GCGCCCAAUGGG_AAAUAAUAAUAAUAUUGCUGGGCUAUUGUCUGGGGC_CAUCU__CCGGCCCCC__A__UU_ ........(((((..(((((((((.....))))))..)))...)))))...........................((((....))))((((..............))))........ (-18.48 = -18.26 + -0.22)

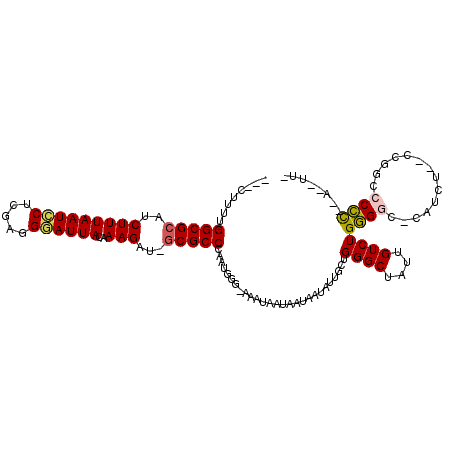

| Location | 7,224,809 – 7,224,917 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.26 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -13.27 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7224809 108 + 27905053 CCAGCAAUAUUAUUAUUAUUU-CCCAAUCG-GCGU--CCUUUGUAAUCCUUCGAGGAUUAAAGGUGCGCCACCAG----CCGUCGUCAACUGAAGUACAACCAAAACGA-AAUCGCA ...((((((....))))((((-(......(-((((--((((..((((((.....)))))))))).))))).....----..(((.((....)).).)).........))-))).)). ( -22.80) >DroVir_CAF1 68835 111 + 1 CCAGCAAUAUUAUUCUUAUUUUCCCAAUGG-GCGCCAUCUUUGUAAUUCCCCGAGAAUUAAAGUGGCGCCAA---CCAACCGUA-AAAACAGAAGUACAACCAAACCAA-AAACAAA ............((((.............(-((((((.(((..((((((.....))))))))))))))))..---......((.-...))))))...............-....... ( -19.60) >DroPse_CAF1 91827 110 + 1 CCAGUAAUAUUAUUAUUAUUU-CCAAUAGG-GCGC-AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG--AACCGUCAACAACAGAAGUACAACCAA-AAUG-AAACGCA ................(((((-(......(-((((-(((((..((((((.....))))))))))))))))....(--(....)).......)))))).......-....-....... ( -25.60) >DroGri_CAF1 92222 109 + 1 CCAGAAAUAUUAUUCUUAUUUUCCCA--GG-GCGCCUUCUUUGUAAUUCCUCGAGGAUUAAAGAGGCGCCAAAAACC-GCCGUA--AAACAGAAGUACAACCAAAC-AA-AAACAAA ...((((((.......))))))....--.(-((((((.(((((....(((....))).)))))))))))).......-......--....................-..-....... ( -22.20) >DroWil_CAF1 84950 107 + 1 CCAGUAAUAUUUUUAUUAUUU-CACAAUUGUGUGC-AUCUUUGUAAUUCUUCGAGAAUUAAAGAUGUGCCG--AU----CCGUA--AAACUGAAGUACAACCAAUAUAAGAAACAAA .((((((((....))))....-.((.((((.(..(-(((((..((((((.....))))))))))))..)))--))----..)).--..))))......................... ( -18.80) >DroPer_CAF1 94906 111 + 1 CCAGUAAUAUUAUUAUUAUUU-CCAAUUGG-GCGC-AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG--AACCGUCAACAACAGAAGUACAACCAAAAAUG-AAACGCA ................(((((-(...((((-((((-(((((..((((((.....))))))))))))))))....(--(....))..)))..))))))............-....... ( -26.40) >consensus CCAGUAAUAUUAUUAUUAUUU_CCCAAUGG_GCGC_AUCUUUGUAAUCCCCCGAGGAUUAAAGAUGCGCCAAAAG___ACCGUA_AAAACAGAAGUACAACCAAAAAAA_AAACAAA .............................(.((((.(((((..((((((.....))))))))))))))))............................................... (-13.27 = -12.50 + -0.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:17 2006