
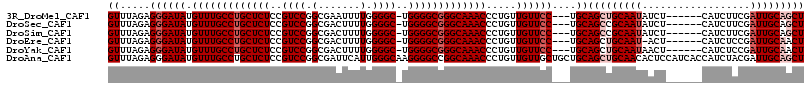
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,219,469 – 7,219,607 |
| Length | 138 |
| Max. P | 0.786178 |

| Location | 7,219,469 – 7,219,575 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -34.59 |
| Energy contribution | -35.75 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7219469 106 + 27905053 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGAAUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAUAUCU------CAUCUUCGAUUGCAGCU ......(((((((.(((((((((((((..((((.(.......).))))-.))))))))))))).....))))))---)..(((((((((....------........))))))))) ( -43.10) >DroSec_CAF1 51347 106 + 1 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCCGCAAUAUCU------CAUCUUCGAUUGCAGCU ......(((((((.(((((((((((((..((((.(.......).))))-.))))))))))))).....))))))---)..(((.(((((....------........))))).))) ( -38.50) >DroSim_CAF1 48017 106 + 1 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCCGCAAUAUCU------CAUCUUCGAUUGCAGCU ......(((((((.(((((((((((((..((((.(.......).))))-.))))))))))))).....))))))---)..(((.(((((....------........))))).))) ( -38.50) >DroEre_CAF1 56458 105 + 1 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAU-ACU------CAUCUCCGAUUGCAACU ......(((((((.(((((((((((((..((((.(.......).))))-.))))))))))))).....))))))---)..((.((((((-.(.------.......))))))).)) ( -37.00) >DroYak_CAF1 56788 106 + 1 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAUAACU------CAUCUCCGAUUGCAACU ......(((((((.(((((((((((((..((((.(.......).))))-.))))))))))))).....))))))---)..((.((((((....------........)))))).)) ( -36.50) >DroAna_CAF1 50446 116 + 1 GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGAUUCAUUGGGCAAGGGGCCGGCAAACCCUGUUGUUGCUGCUGCAGCUGCAACACUCCAUCACCAUCUACGAUUGCAGCU ...(((((((((..(((((((.(((((..((((((.......))))))..))))).)))))))..((((((.((((...)))).))))))....))).)).))))........... ( -41.00) >consensus GUUUAGAGGGAUAUGUUUGCCUGCUCUCCGUCCGGCGACUUUUGGGGC_UGGGGCGGGCAAACCCUGUUGUUCC___UGCAGCUGCAAUAUCU______CAUCUUCGAUUGCAGCU ((.....((((((.(((((((((((((..((((.(.......).))))..))))))))))))).....))))))....))(((((((((..................))))))))) (-34.59 = -35.75 + 1.17)


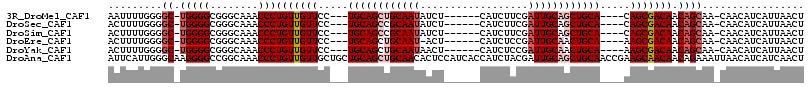
| Location | 7,219,506 – 7,219,607 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -25.41 |
| Energy contribution | -26.44 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7219506 101 + 27905053 AAUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAUAUCU------CAUCUUCGAUUGCAGCUGCA----CAGCGACAACAGCAA-CAACAUCAUUAACU ....(((..((-((....(((....)))(((((((..---((((((((((((....------........))))))))))))----.))))))).))))..-)))........... ( -38.40) >DroSec_CAF1 51384 101 + 1 ACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCCGCAAUAUCU------CAUCUUCGAUUGCAGCUGCA----CAGCGACAACAGCAA-CAACAUCAUUAACU ....(((..((-((....(((....)))(((((((..---((((((.(((((....------........))))).))))))----.))))))).))))..-)))........... ( -33.80) >DroSim_CAF1 48054 101 + 1 ACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCCGCAAUAUCU------CAUCUUCGAUUGCAGCUGCA----CAGCGACAACAGCAA-CAACAUCAUUAACU ....(((..((-((....(((....)))(((((((..---((((((.(((((....------........))))).))))))----.))))))).))))..-)))........... ( -33.80) >DroEre_CAF1 56495 100 + 1 ACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAU-ACU------CAUCUCCGAUUGCAACUGCA----AAGCGACAACAGCAA-CAACAUCAUUAACU ....(((..((-((....(((....)))(((((((..---(((((.((((((-.(.------.......))))))).)))))----.))))))).))))..-)))........... ( -33.10) >DroYak_CAF1 56825 101 + 1 ACUUUUGGGGC-UGGGGCGGGCAAACCCUGUUGUUCC---UGCAGCUGCAAUAACU------CAUCUCCGAUUGCAACUGCA----AAGCGACAACAGCAA-CAACAUCAUUAACU ....(((..((-((....(((....)))(((((((..---(((((.((((((....------........)))))).)))))----.))))))).))))..-)))........... ( -32.60) >DroAna_CAF1 50483 116 + 1 AUUCAUUGGGCAAGGGGCCGGCAAACCCUGUUGUUGCUGCUGCAGCUGCAACACUCCAUCACCAUCUACGAUUGCAGCUGCAACCGAAGCAACAACAGAAAUUAACAUCAUCAACU .....((((((.....)))........(((((((((((..(((((((((((....................))))))))))).....))))))))))).............))).. ( -38.35) >consensus ACUUUUGGGGC_UGGGGCGGGCAAACCCUGUUGUUCC___UGCAGCUGCAAUAUCU______CAUCUUCGAUUGCAGCUGCA____AAGCGACAACAGCAA_CAACAUCAUUAACU .........((.(((((........)))(((((((.....((((((((((((..................)))))))))))).....))))))).))))................. (-25.41 = -26.44 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:11 2006