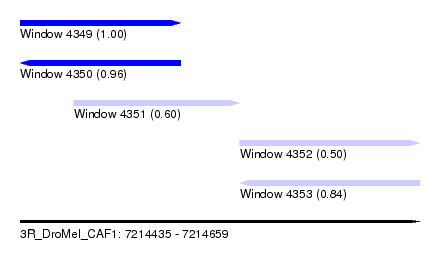
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,214,435 – 7,214,659 |
| Length | 224 |
| Max. P | 0.996254 |

| Location | 7,214,435 – 7,214,525 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 70.16 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -6.24 |
| Energy contribution | -6.63 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.24 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7214435 90 + 27905053 UCCUUAAGUGGGCCCCACCCA--UCCUUGCACACACACACCGAACACCCACAC---UCUGCGGGCCCUCGGCCCCAUUUGU----GUAUGUGUGUGUGU .......(((((.....))))--)....(((((((((((..(......)((((---..((.(((((...)))))))...))----)).))))))))))) ( -34.70) >DroSec_CAF1 46396 84 + 1 UCCUUAAGUGGGCCCCACCCA--CGC--ACACACACACACCGAACACCUCCUC---UCUGCGGCCCCUCUGCCUCAUCUGU----GUG----UGUAUGU .......(((((.....))))--)((--(.(((((((((..((........))---..((.(((......))).))..)))----)))----))).))) ( -26.90) >DroSim_CAF1 43053 84 + 1 UCCUUAAGUGGGCCCCACCCA--CGC--ACACACACACACCGAACACCCCCUC---UCUGCGGCCCCUCUGCCUCAUCUGU----GUU----UGUGUGU .......(((((.....))))--)((--(((((.(((((..((........))---..((.(((......))).))..)))----)).----))))))) ( -26.90) >DroEre_CAF1 51531 86 + 1 UACUUAAGUGGGCCCCACCCA--CGC--A----CACACACCAAACACCCAUUU---ACUCCGGCCCCUCUGAAACAUAUAUAUGUGUG--UGUGUGUGU .......(((((.....))))--)((--(----(((((((.............---....(((.....)))..(((((....))))))--))))))))) ( -25.70) >DroYak_CAF1 51784 82 + 1 UACUUAAGUGGGCCCCACCCA--CGC--G----CACACACCAAACACCCAUUC---ACUCCGUCCCCUCUGCUACAUCUAU----GUG--UGUGUGUGU .......(((((.....))))--)((--(----(((((((((...........---........................)----).)--))))))))) ( -21.45) >DroAna_CAF1 45410 87 + 1 UAUUUAAGGGGGCGCGCCCCCAUCAC--ACACCCACACACCA-ACACAUAUGCAUAAAUUCUUCCACUUGCCACCACACAC----GGG--C---GGUGU .......(((((....))))).....--........(((((.-........(((..............)))..((......----)).--.---))))) ( -20.64) >consensus UACUUAAGUGGGCCCCACCCA__CGC__ACACACACACACCAAACACCCACUC___ACUGCGGCCCCUCUGCCACAUCUAU____GUG__U_UGUGUGU ........((((.....))))............((((((.....................................................)))))). ( -6.24 = -6.63 + 0.39)

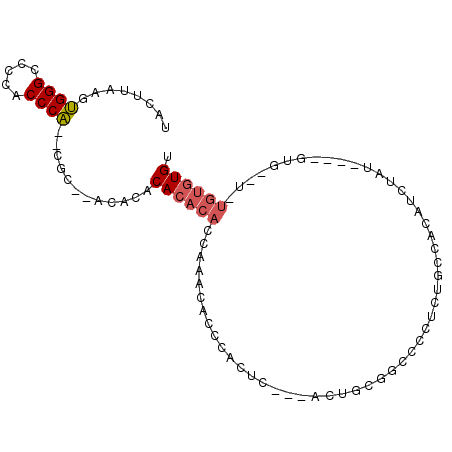
| Location | 7,214,435 – 7,214,525 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.16 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -8.87 |
| Energy contribution | -8.65 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.28 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7214435 90 - 27905053 ACACACACACAUAC----ACAAAUGGGGCCGAGGGCCCGCAGA---GUGUGGGUGUUCGGUGUGUGUGUGCAAGGA--UGGGUGGGGCCCACUUAAGGA .(((((((((...(----(....))..((((((.((((((...---..)))))).)))))))))))))))..(((.--((((.....)))))))..... ( -41.90) >DroSec_CAF1 46396 84 - 1 ACAUACA----CAC----ACAGAUGAGGCAGAGGGGCCGCAGA---GAGGAGGUGUUCGGUGUGUGUGUGU--GCG--UGGGUGGGGCCCACUUAAGGA (((((((----(((----((...((.(((......))).))..---(((......))).))))))))))))--..(--((((.....)))))....... ( -33.40) >DroSim_CAF1 43053 84 - 1 ACACACA----AAC----ACAGAUGAGGCAGAGGGGCCGCAGA---GAGGGGGUGUUCGGUGUGUGUGUGU--GCG--UGGGUGGGGCCCACUUAAGGA (((((((----.((----((...((.(((......))).))..---(((......))).)))).)))))))--..(--((((.....)))))....... ( -29.30) >DroEre_CAF1 51531 86 - 1 ACACACACA--CACACAUAUAUAUGUUUCAGAGGGGCCGGAGU---AAAUGGGUGUUUGGUGUGUG----U--GCG--UGGGUGGGGCCCACUUAAGUA .((((((((--(.((.((((.(((..(((...(....).))).---..))).)))).)))))))))----)--).(--((((.....)))))....... ( -29.10) >DroYak_CAF1 51784 82 - 1 ACACACACA--CAC----AUAGAUGUAGCAGAGGGGACGGAGU---GAAUGGGUGUUUGGUGUGUG----C--GCG--UGGGUGGGGCCCACUUAAGUA .(((((((.--.((----((..((...((...(....)...))---..))..))))...)))))))----.--..(--((((.....)))))....... ( -26.50) >DroAna_CAF1 45410 87 - 1 ACACC---G--CCC----GUGUGUGGUGGCAAGUGGAAGAAUUUAUGCAUAUGUGU-UGGUGUGUGGGUGU--GUGAUGGGGGCGCGCCCCCUUAAAUA .((((---(--(((----..(..(.(..(((.(((.(........).))).)))..-).)..)..))))).--)))..(((((....)))))....... ( -30.40) >consensus ACACACA_A__CAC____ACAGAUGAGGCAGAGGGGCCGCAGA___GAAUGGGUGUUCGGUGUGUGUGUGU__GCG__UGGGUGGGGCCCACUUAAGGA ((((((..........................(....)........(((......))).)))))).............((((.....))))........ ( -8.87 = -8.65 + -0.22)

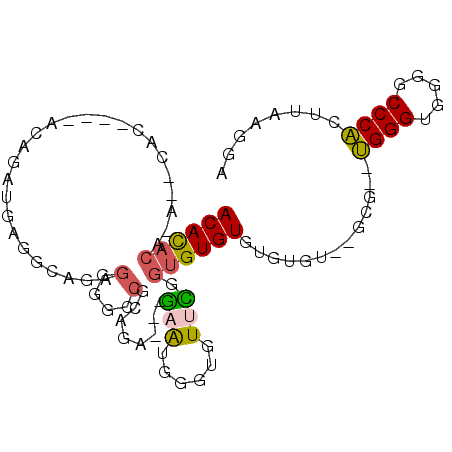
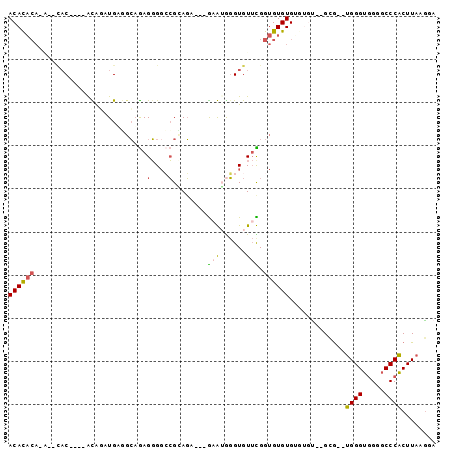
| Location | 7,214,465 – 7,214,558 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -8.60 |
| Energy contribution | -8.96 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7214465 93 + 27905053 ACACACACCGAACACCCACAC---UCUGCGGGCCCUCGGCCCCAUUUGU----GUAUGUGUGUGUGUGUGUUAGUUAAUUAACAAAAGUGCCUUUUGUUC ((((((((...((((..((((---..((.(((((...)))))))...))----))..))))..)))))))).........((((((((...)))))))). ( -34.70) >DroSec_CAF1 46424 89 + 1 ACACACACCGAACACCUCCUC---UCUGCGGCCCCUCUGCCUCAUCUGU----GUG----UGUAUGUGUGUUGGCUAAUUAACAAAAGUGCCUUUUGUUC ((((((((.((..........---..((.(((......))).)))).))----)))----))).................((((((((...)))))))). ( -19.50) >DroSim_CAF1 43081 89 + 1 ACACACACCGAACACCCCCUC---UCUGCGGCCCCUCUGCCUCAUCUGU----GUU----UGUGUGUGUGUUGGCUAAUUAACAAAAGUGCCUUUUGUUC (((((((((((((((......---..((.(((......))).))...))----)))----)).)))))))).........((((((((...)))))))). ( -26.80) >DroEre_CAF1 51556 94 + 1 -CACACACCAAACACCCAUUU---ACUCCGGCCCCUCUGAAACAUAUAUAUGUGUG--UGUGUGUGUGUGUUGGCUAAUUAACAAAAGUGCUUUUUGUUC -(((((((((.((((......---....(((.....)))..((((....)))))))--).)).)))))))..........((((((((...)))))))). ( -18.80) >DroYak_CAF1 51809 90 + 1 -CACACACCAAACACCCAUUC---ACUCCGUCCCCUCUGCUACAUCUAU----GUG--UGUGUGUGUGAGUUGGCUAAUUAACAAAAGUGCCUUUUGUUC -(((((((...((((.(((..---.....((.......)).......))----).)--))))))))))............((((((((...)))))))). ( -17.34) >DroAna_CAF1 45440 90 + 1 CCACACACCA-ACACAUAUGCAUAAAUUCUUCCACUUGCCACCACACAC----GGG--C---GGUGUGUGUUGGCUAAUUAACAAAAGUGGCUUUUGUGC ..........-........(((((((.....((((((((((.(((((((----...--.---.))))))).))))..........))))))..))))))) ( -27.80) >consensus ACACACACCAAACACCCACUC___ACUGCGGCCCCUCUGCCACAUCUAU____GUG__U_UGUGUGUGUGUUGGCUAAUUAACAAAAGUGCCUUUUGUUC .(((((((.......................................................)))))))..........((((((((...)))))))). ( -8.60 = -8.96 + 0.36)


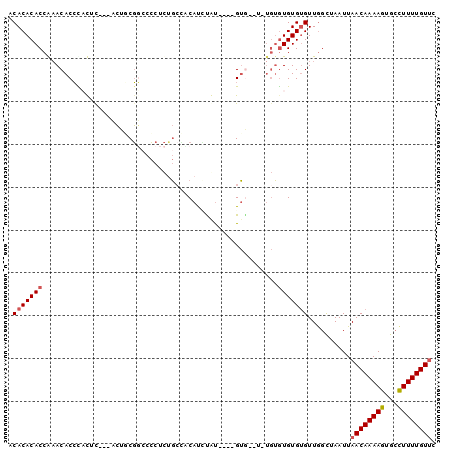
| Location | 7,214,558 – 7,214,659 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -21.75 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7214558 101 + 27905053 GA-------------GGUCCUCAAGCAGGCCACUUAUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGCUCGCCA-AAAUGCAAAUAAUGAAAAGAAAGCGGC-AAAGGACAU ..-------------.(((((.......(((.(((.....((((((.....))))))....((((((..(....((..-....)).....)..))))))))).)))-..))))).. ( -24.50) >DroPse_CAF1 80662 116 + 1 GGAAUGGCAUGGAAUGGUCCUCAAGCAGGCCACUUGUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGUGUGCUUUUGAUGCAAAUAAUGAAAAGAAAGCAGCCAAAGGACAU ....((((......(((.(((.....))))))..(((...((((((.....))))))...(((((((..(...(((.......)))....)..))))))).)))))))........ ( -28.40) >DroSim_CAF1 43170 100 + 1 GA-------------GGUCCUCAAGCAGGCCACUUAUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGCUCGCCA-AAAUGCAAAUAAUGAA-AGAAAGCGGC-AAAGGACAU ..-------------.(((((.......(((.(((.....((((((.....))))))....((((((..(((......-....)))......)))-)))))).)))-..))))).. ( -23.20) >DroYak_CAF1 51899 101 + 1 GA-------------GGUCCUCAAGCAGGCCACUUAUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGCUCGCCA-AAAUGCAAAUAAUGAAAAGAAAGCUGC-AAAGGACAU ..-------------.(((((...((((............((((((.....))))))...(((((((..(....((..-....)).....)..)))))))..))))-..))))).. ( -25.30) >DroAna_CAF1 45530 102 + 1 GA-------------GGUCCUCAAGCCAGCCACUUAUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGCUCGCCA-AAAUGCAAAUAAUGAGAAGAAAGCGGCUAAAGGACAU ..-------------.(((((..((((.((..........((((((.....))))))..((((((....(((......-....)))......))))))...))))))..))))).. ( -29.70) >DroPer_CAF1 84100 116 + 1 GGAAUGGCAUGGAAUGGUCCUCAAGCAGGCCACUUGUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGUGUGCUUUUGAUGCAAAUAAUGAAAAGAAAGCAGCCAAAGGACAU ....((((......(((.(((.....))))))..(((...((((((.....))))))...(((((((..(...(((.......)))....)..))))))).)))))))........ ( -28.40) >consensus GA_____________GGUCCUCAAGCAGGCCACUUAUAUAAUUGCAUUAGCUGCAGUAACUUCUUUUUGUGCUCGCCA_AAAUGCAAAUAAUGAAAAGAAAGCGGC_AAAGGACAU ................(((((...((..((..........((((((.....))))))...(((((((..(....((.......)).....)..))))))).)).))...))))).. (-21.75 = -21.78 + 0.03)

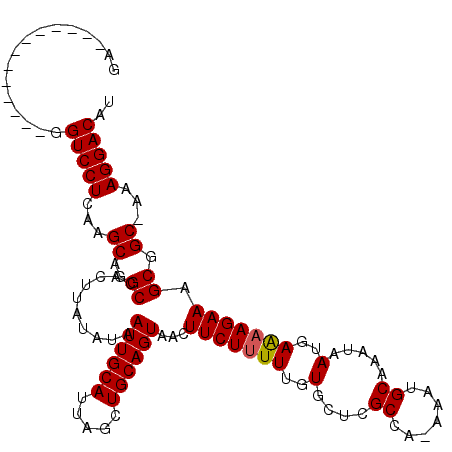
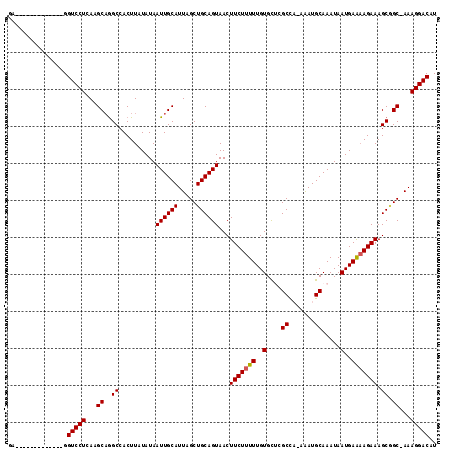
| Location | 7,214,558 – 7,214,659 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.33 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -22.86 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7214558 101 - 27905053 AUGUCCUUU-GCCGCUUUCUUUUCAUUAUUUGCAUUU-UGGCGAGCACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUAUAAGUGGCCUGCUUGAGGACC-------------UC ..((((((.-(((((((((((((.....(((((....-..)))))....))))))).....((((.....))))........))))))......)))))).-------------.. ( -25.90) >DroPse_CAF1 80662 116 - 1 AUGUCCUUUGGCUGCUUUCUUUUCAUUAUUUGCAUCAAAAGCACACACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUACAAGUGGCCUGCUUGAGGACCAUUCCAUGCCAUUCC ..(((((((((((((((((((((.......(((.......)))......)))))))).....))))))))..........(((((.....))))))))))................ ( -26.42) >DroSim_CAF1 43170 100 - 1 AUGUCCUUU-GCCGCUUUCU-UUCAUUAUUUGCAUUU-UGGCGAGCACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUAUAAGUGGCCUGCUUGAGGACC-------------UC ..((((((.-(((((((...-.....((.((((((..-((((..(((..............))).)))))))))).))...)))))))......)))))).-------------.. ( -26.14) >DroYak_CAF1 51899 101 - 1 AUGUCCUUU-GCAGCUUUCUUUUCAUUAUUUGCAUUU-UGGCGAGCACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUAUAAGUGGCCUGCUUGAGGACC-------------UC ..((((((.-((((((..(((.....((.((((((..-((((..(((..............))).)))))))))).))...))).)).))))..)))))).-------------.. ( -27.34) >DroAna_CAF1 45530 102 - 1 AUGUCCUUUAGCCGCUUUCUUCUCAUUAUUUGCAUUU-UGGCGAGCACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUAUAAGUGGCUGGCUUGAGGACC-------------UC ..((((((.(((((((..(((.....((.((((((..-((((..(((..............))).)))))))))).))...))).))).)))).)))))).-------------.. ( -30.54) >DroPer_CAF1 84100 116 - 1 AUGUCCUUUGGCUGCUUUCUUUUCAUUAUUUGCAUCAAAAGCACACACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUACAAGUGGCCUGCUUGAGGACCAUUCCAUGCCAUUCC ..(((((((((((((((((((((.......(((.......)))......)))))))).....))))))))..........(((((.....))))))))))................ ( -26.42) >consensus AUGUCCUUU_GCCGCUUUCUUUUCAUUAUUUGCAUUU_UGGCGAGCACAAAAAGAAGUUACUGCAGCUAAUGCAAUUAUAUAAGUGGCCUGCUUGAGGACC_____________UC ..((((((..(((((((((((((.....(((((.......)))))....))))))).....((((.....))))........))))))......))))))................ (-22.86 = -23.08 + 0.22)

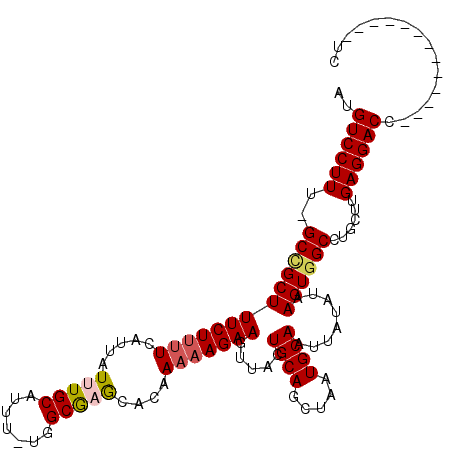
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:06 2006