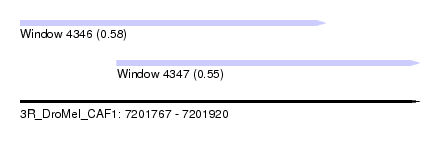
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,201,767 – 7,201,920 |
| Length | 153 |
| Max. P | 0.583806 |

| Location | 7,201,767 – 7,201,884 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -21.97 |
| Energy contribution | -23.00 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
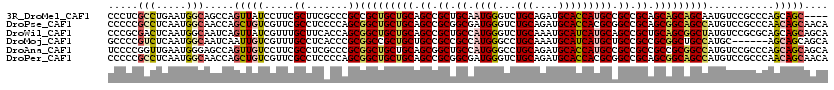
>3R_DroMel_CAF1 7201767 117 + 27905053 -CUUUA--CUGCCGCCCAACUGCCAACAGUUGAUCUCCGCCCCUCGCCUGAAUGGCAGCCAGUUAUCCUUCGCUUCGCCCGCCGCUGCUGCAGCCGCUGCAAUGGGUCUGCAGAUGCACC -.....--((((.(((((.((((((.(((.(((..........))).)))..))))))..................((..((.((((...)))).)).))..)))))..))))....... ( -32.90) >DroPse_CAF1 77088 120 + 1 UCCAUGCCAUGCCCCCCAAUUGCCAGCAGCUGAUAGCCGCCCCCCGCCUCAAUGGCAACCAGCUGUCGUUCGCCUCCCCAGCGGCUGCUGCAGCCGCGGCGAUGGGUCUGCAGAUGCACC ....(((..(((..((((.(((((.((.((((.((((.(((....(((.....))).....((((..(.......)..))))))).)))))))).)))))))))))...)))...))).. ( -49.60) >DroGri_CAF1 76941 108 + 1 ------------UGCCCAAUUGCCAGCAGCUAAUUGCUGCACCACGUUUAAAUGGCAAUCAGCUGUCUUUUGCCUCACCCGCCGCAGCUGCUGCCGCCGCGAUGGGUCUGCAAAUGCACC ------------.(((((.((((..(((((.....))))).............((((..(((((((.....((.......)).))))))).))))...))))))))).(((....))).. ( -37.50) >DroWil_CAF1 68709 117 + 1 -CCUUG--GCCAUGGCCAAUUGUCAGCAAUUGAUUGCUGCCCCGCGACUCAAUGGCAAUCAGUUAUCGUUUGCUUCACCAGCGGCUGCUGCAGCCGCUGCCAUGGGUCUGCAAAUGCAUC -..(((--((....))))).(((((((((....))))))....((((((((.((((....(((........))).....((((((((...)))))))))))))))))).))....))).. ( -43.90) >DroAna_CAF1 41863 117 + 1 -CGUUG--AUGCCGCCCAACUGUCAGCAGCUAAUAGCUGCUCCCCGGUUGAAUGGGAGCCAGUUGUCCUUCGCCUCGCCCGCGGCUGCUGCAGCGGCUGCCAUGGGCCUGCAGAUGCACC -.((..--.(((.(((((((((..((((((.....))))))...)))))..((((.((((.(((((.....(((.(....).)))....))))))))).))))))))..)))...))... ( -49.20) >DroPer_CAF1 80526 120 + 1 UCCAUGCCAUGCCCCCCAAUUGCCAGCAGCUGAUAGCCGCCCCCCGCCUCAAUGGCAACCAGCUGUCGUUCGCCUCCCCAGCGGCUGCUGCAGCCGCGGCGAUGGGUCUGCAGAUGCACC ....(((..(((..((((.(((((.((.((((.((((.(((....(((.....))).....((((..(.......)..))))))).)))))))).)))))))))))...)))...))).. ( -49.60) >consensus _CCUUG__AUGCCGCCCAAUUGCCAGCAGCUGAUAGCCGCCCCCCGCCUCAAUGGCAACCAGCUGUCGUUCGCCUCACCAGCGGCUGCUGCAGCCGCUGCGAUGGGUCUGCAGAUGCACC .............(((((.......(((((....((((((.....(((.....))).....((........)).......))))))))))).((....))..))))).(((....))).. (-21.97 = -23.00 + 1.03)

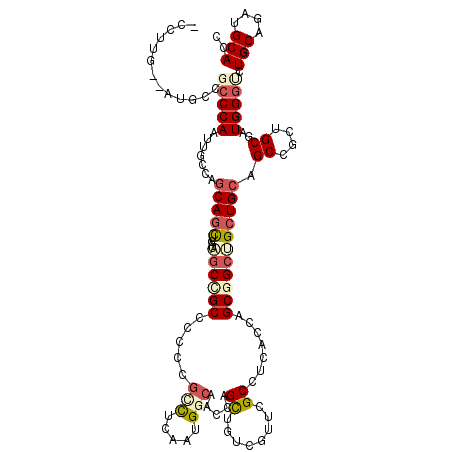
| Location | 7,201,804 – 7,201,920 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -48.53 |
| Consensus MFE | -35.38 |
| Energy contribution | -35.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7201804 116 + 27905053 CCCUCGCCUGAAUGGCAGCCAGUUAUCCUUCGCUUCGCCCGCCGCUGCUGCAGCCGCUGCAAUGGGUCUGCAGAUGCACCAUGCCGCCGCAGCAGCAGCAAUGUCCGCCCAGCAGC---- .....(((.....)))...............(((..((..((.((((((((.((.((.(((.(((...(((....))))))))).)).)).))))))))...))..))..)))...---- ( -41.10) >DroPse_CAF1 77128 120 + 1 CCCCCGCCUCAAUGGCAACCAGCUGUCGUUCGCCUCCCCAGCGGCUGCUGCAGCCGCGGCGAUGGGUCUGCAGAUGCACCACGCGGCCGCAGCGGCAGCCAUGUCCGCCCAACAGCAACA .....(((.....))).....(((((.(..(((.......))(((((((((.((.((.(((.(((...(((....))))))))).)).)).)))))))))......)..).))))).... ( -51.10) >DroWil_CAF1 68746 120 + 1 CCCGCGACUCAAUGGCAAUCAGUUAUCGUUUGCUUCACCAGCGGCUGCUGCAGCCGCUGCCAUGGGUCUGCAAAUGCAUCAUGCAGCCGCUGCAGCGGCUAUGUCCGCGCAGCAGCAGCA ...((((((((.((((....(((........))).....((((((((...))))))))))))))))))(((...(((.....)))...(((((.((((......))))))))).))))). ( -52.30) >DroMoj_CAF1 73153 114 + 1 GCCCCGUCUCAAUGGCAAUCAAUUGUCGUUUGCCUCACCCGCGGCCGCUGCUGCCGCCGCCAUGGGCCUGCAAAUGCAUCAUGCUGCCGCCGCGGCUGCCAUGC------AGCAGCAGCA .............((((......((.(((((((....((((((((.((....)).)))))...)))...)))))))))......))))((.((.(((((...))------))).)).)). ( -44.80) >DroAna_CAF1 41900 120 + 1 UCCCCGGUUGAAUGGGAGCCAGUUGUCCUUCGCCUCGCCCGCGGCUGCUGCAGCGGCUGCCAUGGGCCUGCAGAUGCACCAUGCCGCCGCCGCCGCGGCCAUGUCCGCCCAGCAGCAGCA ...((((.(((..((((........)))).....))).))).)((((((((.(((((.(((((((((........)).)))))..)).))))).((((......))))...)))))))). ( -50.80) >DroPer_CAF1 80566 120 + 1 CCCCCGCCUCAAUGGCAACCAGCUGUCGUUCGCCUCCCCAGCGGCUGCUGCAGCCGCGGCGAUGGGUCUGCAGAUGCACCACGCGGCCGCAGCGGCAGCCAUGUCCGCCCAACAGCAACA .....(((.....))).....(((((.(..(((.......))(((((((((.((.((.(((.(((...(((....))))))))).)).)).)))))))))......)..).))))).... ( -51.10) >consensus CCCCCGCCUCAAUGGCAACCAGUUGUCGUUCGCCUCACCAGCGGCUGCUGCAGCCGCUGCCAUGGGUCUGCAGAUGCACCAUGCCGCCGCAGCGGCAGCCAUGUCCGCCCAGCAGCAGCA .....(((.....))).....(((((.....((.......))(((((((((.((.((.((.((((...(((....))))))))).)).)).)))))))))...........))))).... (-35.38 = -35.88 + 0.50)
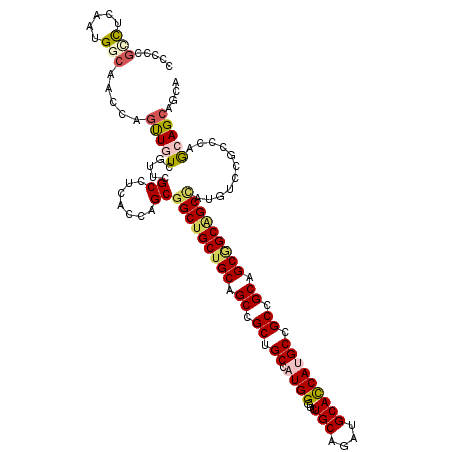
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:01 2006