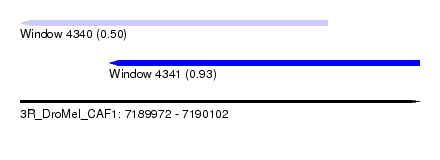
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,189,972 – 7,190,102 |
| Length | 130 |
| Max. P | 0.933319 |

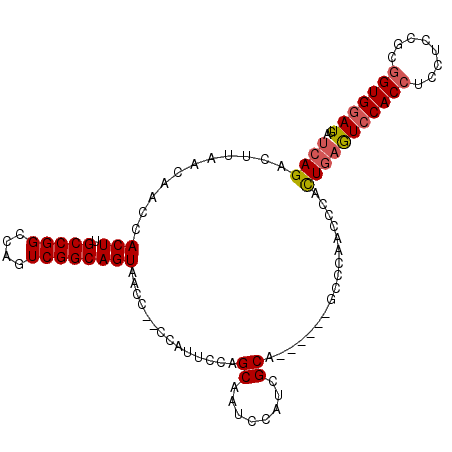
| Location | 7,189,972 – 7,190,072 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 91.80 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -21.66 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7189972 100 - 27905053 GGCAGUAACCCCCCAUGCCAGCAAUCCAACGCA------GCCCAACCCAUUGAGUCCACCUCCUCCGCGGUGGAUGACGCUUCUGACAGCUCGUCAGCUGGCAUCA ..............((((((((........(.(------((.(((....))).(((((((........)))))))...))).)((((.....)))))))))))).. ( -30.00) >DroSec_CAF1 30717 98 - 1 GGCAGUAACC--CCAUUCCAGCAAUCCAUCGCU------GCCCAACCCACUAUGUCCACCUCCUCCGCGGUGGAUGACUCUUCUGACAGCUCGUCAGCUGGCAUCA ((((((....--..................)))------)))...........(((((((........)))))))((..((.(((((.....)))))..))..)). ( -25.15) >DroSim_CAF1 30719 98 - 1 GGCAGUAACC--CCAUUCCAGCAAUCCAUCGCU------GCCCAACCCACUGAGUCCACCUCCUCCGCGGUGGAUGACUCUUCUGACAGCUCGUCAGCUGGCAUCA ((((((....--..................)))------)))...........(((((((........)))))))((..((.(((((.....)))))..))..)). ( -25.25) >DroEre_CAF1 36169 98 - 1 GGCAGUAACC--CCAUACCAGCAAUCCAUCGCA------GCCCAACCCACUGAAUACACCUCCUCCGCGGUGGAUGACUCUUCUGACAGCUCGUCAGCUGGCAUCA ((........--))...(((((.(((((((((.------...........................)))))))))........((((.....)))))))))..... ( -24.19) >DroYak_CAF1 36289 104 - 1 GGCAGUAACC--CCAUUCCAGCAAUCCAUCGCCGCAGUGGCCCAACCCACUGAAUCCACCUCCUCCGCUGUGGAUGACGCUUCUGACAGCUCGUCAGCUGGCAUCA ..........--.(((((((((............((((((......))))))..............)))).)))))..(((.(((((.....)))))..))).... ( -28.27) >consensus GGCAGUAACC__CCAUUCCAGCAAUCCAUCGCA______GCCCAACCCACUGAGUCCACCUCCUCCGCGGUGGAUGACUCUUCUGACAGCUCGUCAGCUGGCAUCA .................(((((.(((((((((..................................)))))))))........((((.....)))))))))..... (-21.66 = -22.06 + 0.40)

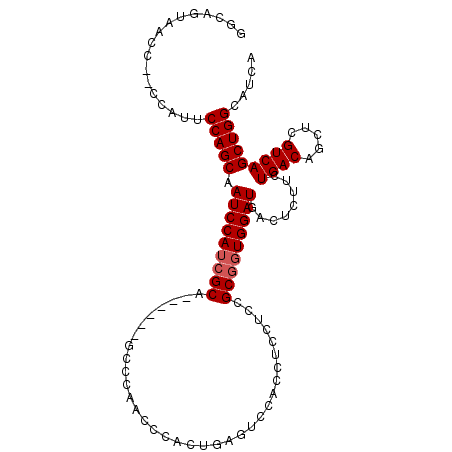

| Location | 7,190,001 – 7,190,102 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7190001 101 - 27905053 UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACCCCCCAUGCCAGCAAUCCAACGCA------GCCCAACCCAUUGAGUCCACCUCCUCCGCGGUGGAUGA ((((...........(((.(((((....))))))))..........((..((........)).------))........))))(((((((........))))))).. ( -23.40) >DroSec_CAF1 30746 99 - 1 UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACC--CCAUUCCAGCAAUCCAUCGCU------GCCCAACCCACUAUGUCCACCUCCUCCGCGGUGGAUGA ...............(((.(((((....))))))))....--......((((........)))------).............(((((((........))))))).. ( -24.70) >DroSim_CAF1 30748 99 - 1 UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACC--CCAUUCCAGCAAUCCAUCGCU------GCCCAACCCACUGAGUCCACCUCCUCCGCGGUGGAUGA ((((...........(((.(((((....))))))))....--......((((........)))------).........))))(((((((........))))))).. ( -28.70) >DroEre_CAF1 36198 99 - 1 UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACC--CCAUACCAGCAAUCCAUCGCA------GCCCAACCCACUGAAUACACCUCCUCCGCGGUGGAUGA ...............(((.(((((....))))))))....--...........(((((((((.------...........................))))))))).. ( -21.59) >DroYak_CAF1 36318 105 - 1 UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACC--CCAUUCCAGCAAUCCAUCGCCGCAGUGGCCCAACCCACUGAAUCCACCUCCUCCGCUGUGGAUGA ...............(((.(((((....))))))))....--.(((((((((............((((((......))))))..............)))).))))). ( -27.77) >consensus UCAGACUUAACAACCACUUGCCGGCCAGUCGGCAGUAACC__CCAUUCCAGCAAUCCAUCGCA______GCCCAACCCACUGAGUCCACCUCCUCCGCGGUGGAUGA ((((...........(((.(((((....))))))))..............((........)).................))))(((((((........))))))).. (-21.14 = -21.54 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:55 2006