
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,184,160 – 7,184,254 |
| Length | 94 |
| Max. P | 0.962889 |

| Location | 7,184,160 – 7,184,254 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.64 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.33 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
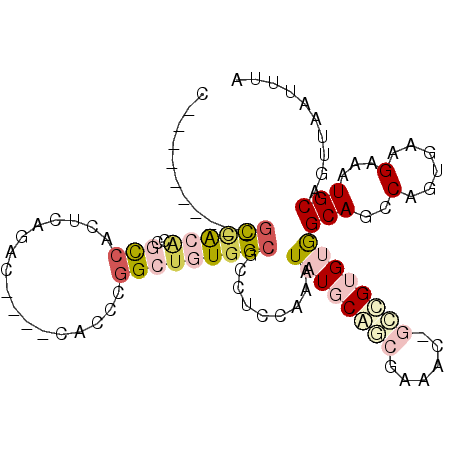
>3R_DroMel_CAF1 7184160 94 + 27905053 C----------CCUACACCGCCACUCAGAC----CACCUGGCUGUGGCGCCUCCAAAUAUGCGGCGCAACAGCCGUGUGGCAGGCUGUGAAGAAAUGCAGUUAAUUUA .----------.((.(((.(((.....(.(----(((.(((((((.(((((.(.......).))))).))))))).))))).))).))).))................ ( -35.90) >DroPse_CAF1 60287 101 + 1 GA-AGCAGGCAGCCAGGCAGCCAGGCAGGCCCUUCAGCCGGCUGUGGCGCCUCCAAAUAUGCAACGAG------GAGCAGCAGCCACUGAAGAAAUGCAGUUAAUUUA ..-.((((((.(((.((...)).)))..)))((((((..((((((.((.((((............)))------).)).)))))).))))))...))).......... ( -39.80) >DroSim_CAF1 24969 94 + 1 C----------GCCACACCACCACUCAGAC----CACCUGGCUGUGGCGCCUCCAAAUAUGCGGCGCAACAGCCGUGUGGCAGGCUGUGAAGAAAUGCAGUUAAUUUA .----------(((((((........((..----...))((((((.(((((.(.......).))))).))))))))))))).((((((........))))))...... ( -38.70) >DroYak_CAF1 25624 99 + 1 C-----CCUUGGCCACCCCACCACUCAGAC----CACCUGGCUGUGGCGCCUCCAAAUAUGCGGCGAAACGGCCGUGUGGCAGGCUGUGAAGAAAUGCAGUUAAUUUA .-----...(((........)))....(.(----(((.(((((((..((((.(.......).))))..))))))).))))).((((((........))))))...... ( -32.50) >DroAna_CAF1 24146 88 + 1 U----------GUGGCAUGGCAAGUU----------GCCGGCUGUGGCGCCUCCAAAUAUGCACCGAGAGCGGUGUGUGGCAGCCAGUGAAGAAAUGCAGUUAAUUUA .----------...(((((((.....----------)))((((((((.....))...(((((((((....))))))))))))))).........)))).......... ( -31.00) >DroPer_CAF1 62063 102 + 1 GAAAGCAGGCAGCCAGGCAGUCAGGCAGGCCCUUCAGCCGGCUGUGGCGCCUCCAAAUAUGCAACGAG------GAGCAGCAGCCACUGAAGAAAUGCAGUUAAUUUA ....((((((.(((.........)))..)))((((((..((((((.((.((((............)))------).)).)))))).))))))...))).......... ( -38.40) >consensus C__________GCCACACCGCCACUCAGAC____CACCCGGCUGUGGCGCCUCCAAAUAUGCAGCGAAAC_GCCGUGUGGCAGCCAGUGAAGAAAUGCAGUUAAUUUA ...........((((((..(((.................))))))))).........((((((((......))))))))(((..(......)...))).......... (-11.98 = -13.73 + 1.75)


| Location | 7,184,160 – 7,184,254 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.64 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -11.23 |
| Energy contribution | -10.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
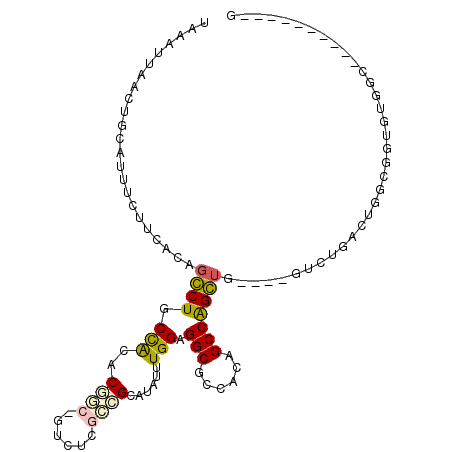
>3R_DroMel_CAF1 7184160 94 - 27905053 UAAAUUAACUGCAUUUCUUCACAGCCUGCCACACGGCUGUUGCGCCGCAUAUUUGGAGGCGCCACAGCCAGGUG----GUCUGAGUGGCGGUGUAGG----------G ........((((((..((.(.(((...(((((..((((((.(((((.((....))..))))).))))))..)))----))))).).))..)))))).----------. ( -42.70) >DroPse_CAF1 60287 101 - 1 UAAAUUAACUGCAUUUCUUCAGUGGCUGCUGCUC------CUCGUUGCAUAUUUGGAGGCGCCACAGCCGGCUGAAGGGCCUGCCUGGCUGCCUGGCUGCCUGCU-UC ..........(((...((((((((((((..((.(------(((............)))).))..))))).)))))))(((..(((.((...)).))).)))))).-.. ( -39.70) >DroSim_CAF1 24969 94 - 1 UAAAUUAACUGCAUUUCUUCACAGCCUGCCACACGGCUGUUGCGCCGCAUAUUUGGAGGCGCCACAGCCAGGUG----GUCUGAGUGGUGGUGUGGC----------G ........(..(((..((.(.(((...(((((..((((((.(((((.((....))..))))).))))))..)))----))))).).))..)))..).----------. ( -40.60) >DroYak_CAF1 25624 99 - 1 UAAAUUAACUGCAUUUCUUCACAGCCUGCCACACGGCCGUUUCGCCGCAUAUUUGGAGGCGCCACAGCCAGGUG----GUCUGAGUGGUGGGGUGGCCAAGG-----G ..........((....((((((.(((.(((((..(((.((..((((.((....))..))))..)).)))..)))----))..).)).))))))..)).....-----. ( -32.20) >DroAna_CAF1 24146 88 - 1 UAAAUUAACUGCAUUUCUUCACUGGCUGCCACACACCGCUCUCGGUGCAUAUUUGGAGGCGCCACAGCCGGC----------AACUUGCCAUGCCAC----------A ..........((((........((((.(((...(((((....)))))((....))..))))))).....(((----------.....)))))))...----------. ( -24.00) >DroPer_CAF1 62063 102 - 1 UAAAUUAACUGCAUUUCUUCAGUGGCUGCUGCUC------CUCGUUGCAUAUUUGGAGGCGCCACAGCCGGCUGAAGGGCCUGCCUGACUGCCUGGCUGCCUGCUUUC ..........(((...((((((((((((..((.(------(((............)))).))..))))).)))))))(((..(((.........))).)))))).... ( -38.10) >consensus UAAAUUAACUGCAUUUCUUCACAGCCUGCCACACGGC_GUCUCGCCGCAUAUUUGGAGGCGCCACAGCCAGCUG____GUCUGACUGGCGGUGUGGC__________G .......................((((.(((..((((......))))......))).(((......)))))))................................... (-11.23 = -10.87 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:53 2006