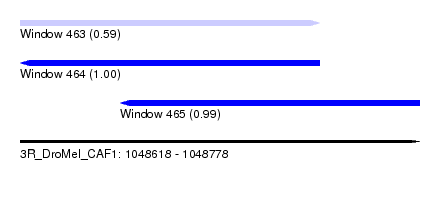
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,048,618 – 1,048,778 |
| Length | 160 |
| Max. P | 0.996533 |

| Location | 1,048,618 – 1,048,738 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -43.23 |
| Consensus MFE | -40.00 |
| Energy contribution | -40.80 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
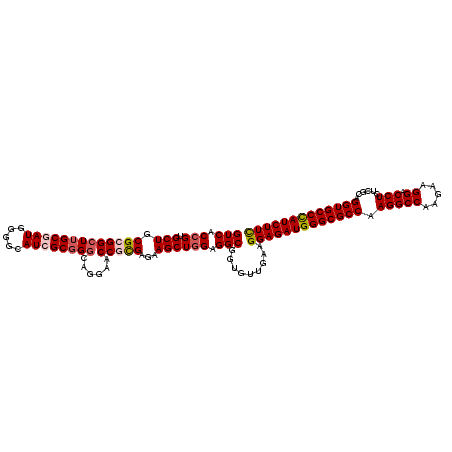
>3R_DroMel_CAF1 1048618 120 + 27905053 CCAUCUCAAGCAGGGAGGCCUGGUCGUUGACGUCCGCGAUGAAGAUGGGCACCUGCGAGAGGUCCUUCUUGGCCUUGGCGCCCAUCUCCUUCAACACCGCCUCCAGCUUCUCGCGGUUCC ((.....((((..((((((..(((.(((((((((...)))).((((((((.((.....((((((......)))))))).))))))))...)))))))))))))).)))).....)).... ( -48.20) >DroSec_CAF1 20531 120 + 1 CCAUCUCCAGCAGGGAGGUCUGGUCGUUGACGUCCGCGAUGAAGAUGGGCACCUGCGAGAGGUCCUUCUUGGCCUUGGCGCCCAUCUCCUUCAACACCGCCUCCAGCUUCUCGCGGUUCC ((......(((..((((((..(((.(((((((((...)))).((((((((.((.....((((((......)))))))).))))))))...)))))))))))))).)))......)).... ( -45.70) >DroSim_CAF1 20411 120 + 1 CCAUCUCCAGCAGGGAGGCCUGGUCGUUGACGUCCGCGAUGAAGAUGGGCACCUGCGAGAGGUCCUUCUUGGCCUUGGCGCCCAUCUCCUUCAACACCGCCUCCAGCUUCUCGCGGUUCC ((......(((..((((((..(((.(((((((((...)))).((((((((.((.....((((((......)))))))).))))))))...)))))))))))))).)))......)).... ( -47.10) >DroEre_CAF1 20689 120 + 1 CCAUCUCCAGCAGGGAGGCCUCGUCGUUGACGUCCGCGAUGAAGAUAGGCACCUGCGACAGGUCCUUUUUGGCCUUGGCGCCCAUCUCCUUCAACACCGCCUCCAGCUUCUCGCGGUUUC ((......(((..((((((.(((((((.(....).)))))))((((.(((.((......(((((......))))).)).))).))))...........)))))).)))......)).... ( -38.60) >DroYak_CAF1 21448 120 + 1 CCAUCUCCAGCAGGGAAGUCUCGUCGUUGACGUCCGCGAUAAAGAUGGGCACCUGUGAGAGGUCCUUCUUGGCCUUGGCGCCCAUCUCCUUUAAAACCGCCUCCAGCUUCUCACGGUUUC ((((((...((..(((.(((........))).))))).....))))))....(((((((((((......(((....((((.................)))).)))))))))))))).... ( -36.53) >consensus CCAUCUCCAGCAGGGAGGCCUGGUCGUUGACGUCCGCGAUGAAGAUGGGCACCUGCGAGAGGUCCUUCUUGGCCUUGGCGCCCAUCUCCUUCAACACCGCCUCCAGCUUCUCGCGGUUCC ((......(((..((((((..(((.(((((((....))....((((((((.((.....((((((......)))))))).))))))))...)))))))))))))).)))......)).... (-40.00 = -40.80 + 0.80)

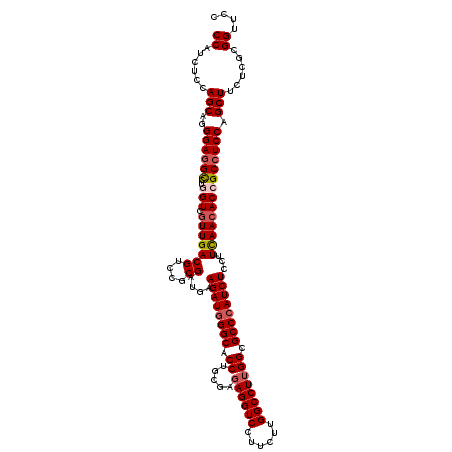
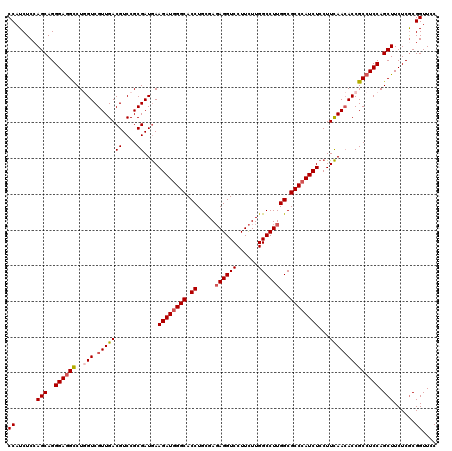
| Location | 1,048,618 – 1,048,738 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -50.52 |
| Consensus MFE | -46.06 |
| Energy contribution | -45.38 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1048618 120 - 27905053 GGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUCAUCGCGGACGUCAACGACCAGGCCUCCCUGCUUGAGAUGG ....((((...((((.((((((((((((((.(((((((((((((.(((((.....)).)))......)))))))))))))..((....))))))).)))..))))))..))))..).))) ( -53.10) >DroSec_CAF1 20531 120 - 1 GGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUCAUCGCGGACGUCAACGACCAGACCUCCCUGCUGGAGAUGG ....((((....(((.((((((((((((((.(((((((((((((.(((((.....)).)))......)))))))))))))..((....))))))).))).).)))))..)))...).))) ( -50.50) >DroSim_CAF1 20411 120 - 1 GGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUCAUCGCGGACGUCAACGACCAGGCCUCCCUGCUGGAGAUGG ....((((....(((.((((((((((((((.(((((((((((((.(((((.....)).)))......)))))))))))))..((....))))))).)))..))))))..)))...).))) ( -52.90) >DroEre_CAF1 20689 120 - 1 GAAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAAAAGGACCUGUCGCAGGUGCCUAUCUUCAUCGCGGACGUCAACGACGAGGCCUCCCUGCUGGAGAUGG ....((((....(((.((((((..(((((..(((((((((((((.(((((.....)).)))......)))))))))))))...((....))...)))))..))))))..)))...).))) ( -50.20) >DroYak_CAF1 21448 120 - 1 GAAACCGUGAGAAGCUGGAGGCGGUUUUAAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCACAGGUGCCCAUCUUUAUCGCGGACGUCAACGACGAGACUUCCCUGCUGGAGAUGG ....(((((((((((((....)))))))...(((((((((((((.(((((.....)).)))......))))))))))))).)))))).((((...))))...(.((((....)).)).). ( -45.90) >consensus GGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUCAUCGCGGACGUCAACGACCAGGCCUCCCUGCUGGAGAUGG ....((((((...((((....))))......(((((((((((((.(((((.....)).)))......))))))))))))).))))))..(((........)))((((.....)))).... (-46.06 = -45.38 + -0.68)

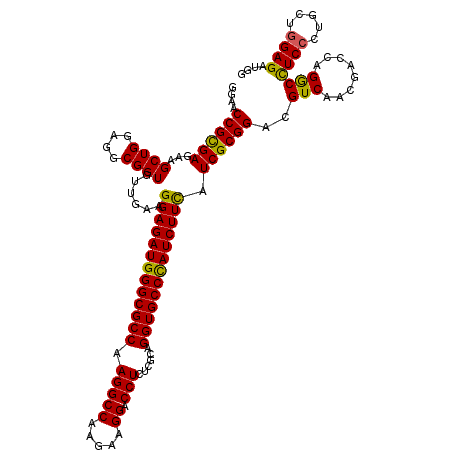
| Location | 1,048,658 – 1,048,778 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -52.00 |
| Consensus MFE | -47.66 |
| Energy contribution | -48.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1048658 120 - 27905053 GUCACGGUGCUGCGCGGCUUGCGAUGGGGCAUCGCUGGCAGGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUC ....(((..((((...(((.((((((...)))))).))).....((((.....)).))..))))..)))..(((((((((((((.(((((.....)).)))......))))))))))))) ( -53.00) >DroSec_CAF1 20571 120 - 1 GUCACCGUGCUGCGCGGCUUGCGAUGGGGCAUAGCGGGCAGGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUC ..((((((.((((.(.((((((......))).))).)))))...((((.....)).))..)))))).....(((((((((((((.(((((.....)).)))......))))))))))))) ( -53.70) >DroSim_CAF1 20451 120 - 1 GUCACCGUGCUGCGCGGCUUGCGAUGGGGCAUAGCGGGCAGGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUC ..((((((.((((.(.((((((......))).))).)))))...((((.....)).))..)))))).....(((((((((((((.(((((.....)).)))......))))))))))))) ( -53.70) >DroEre_CAF1 20729 120 - 1 GUCACCGUGCUGCGAGGCUUGCGAUGGGGAAUCGCGGGCAGAAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAAAAGGACCUGUCGCAGGUGCCUAUCUUC ..((((((.((.((..(((((((((.....))))))))).......((.....)))).)))))))).....(((((((((((((.(((((.....)).)))......))))))))))))) ( -52.40) >DroYak_CAF1 21488 120 - 1 GUCACCGUGCUGCGAGGGUUGCGAUGGGGAAUCGCGGGCAGAAACCGUGAGAAGCUGGAGGCGGUUUUAAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCACAGGUGCCCAUCUUU .((.((((((.((....)).)).)))).)).((((((.......))))))(((((((....)))))))...(((((((((((((.(((((.....)).)))......))))))))))))) ( -47.20) >consensus GUCACCGUGCUGCGCGGCUUGCGAUGGGGCAUCGCGGGCAGGAACCGCGAGAAGCUGGAGGCGGUGUUGAAGGAGAUGGGCGCCAAGGCCAAGAAGGACCUCUCGCAGGUGCCCAUCUUC (((.(((.(((.(((((((((((((.....))))))))......)))))...)))))).))).........(((((((((((((.(((((.....)).)))......))))))))))))) (-47.66 = -48.58 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:06 2006