| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
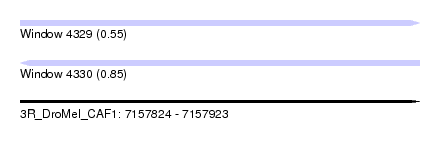
| Location | 7,157,824 – 7,157,923 |
| Length | 99 |
| Max. P | 0.853950 |

| Location | 7,157,824 – 7,157,923 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -26.98 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
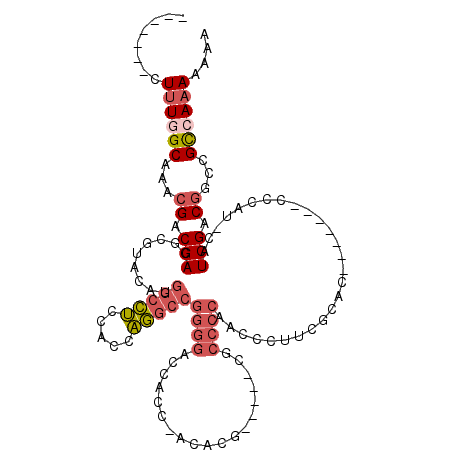
>3R_DroMel_CAF1 7157824 99 + 27905053 UUUUUUUGGCGGCCGUCGAUG-AUGGG-------GUGCGAAGGGUUGGGGCG-----CGUGG-UGUGUUCCCCGGCCUGGUGGAGGCCAUGUACGCGUCGUCGUUUGCUAAAG------- ....((((((((....(((((-(((.(-------.((((..(((((((((.(-----((...-..))).)))))))))(((....))).))))).)))))))).)))))))).------- ( -41.20) >DroSec_CAF1 90810 99 + 1 UUUUUUUGACGGCCGUCGAUG-GUGGC-------GUGCGAAGGGUUGGGGUG-----CGUGG-GGUGGUCCCCGGCCUGGUGGAGGCCAUGUACGCGUCGUCGUUUGCCAAAG------- ..........(((...(((((-..(.(-------(((((..(((((((((.(-----(....-....)))))))))))(((....))).)))))))..)))))...)))....------- ( -41.10) >DroSim_CAF1 103984 99 + 1 UUUUUUUGGCGGCCGUCGAUG-AUGGG-------GUGCGAAGGGUUGGGGUG-----CGUGG-GGUGGUCCCCGGCCUGGCGGAGGCCAUGUACGCGUCGUCGUUUGCCAAAG------- ....((((((((....(((((-(((.(-------.((((..(((((((((.(-----(....-....)))))))))))(((....))).))))).)))))))).)))))))).------- ( -44.30) >DroEre_CAF1 92524 99 + 1 UUUUUUUGGCGGCCGUCGAUG-AUGGG-------GUGCGAAGGGAUGGGGCG-----UGUAU-GGUGGUCCCCGGCCUGGUGGAGGCCAUGUACGCGUCGUCGUUUGCCAAAG------- ....((((((..(((((...)-)))).-------.....(((.(((((.(((-----(((((-(((..(((((.....)).))).))))))))))).))))).))))))))).------- ( -41.30) >DroYak_CAF1 94596 104 + 1 UUU-UUUGGCGGCCGUCGAUGAAUGGG-------GUGCGAAGGGAUGGGGCGUGGGGUGUGU-GGUGGUCCCCGGCCUGGUGGAGGCCAUGUACGCGUCGUCGUUUGCCAAAG------- ...-((((((.(((.(((.....))).-------).)).(((.(((((.((((((((.....-......))))(((((.....)))))....)))).))))).))))))))).------- ( -41.20) >DroAna_CAF1 99768 106 + 1 UUU-UUUGGCGGUUGUCGAUG-GAGGUGGACUCUGGGCGAUGGGUUAUGGGA-----AGGAUGGGUGGU-------UAGGUGCCAACCAUGUACGCGUCGUCGUUUGCUAGAGCCGUUCC ...-...(((((((.(.(..(-(((.....)))).(((((((.(((((((..-----........((((-------.....)))).))))).)).))))))).....).).))))))).. ( -32.60) >consensus UUUUUUUGGCGGCCGUCGAUG_AUGGG_______GUGCGAAGGGUUGGGGCG_____CGUGG_GGUGGUCCCCGGCCUGGUGGAGGCCAUGUACGCGUCGUCGUUUGCCAAAG_______ ....((((((((.((.(((((.............(((((..(((..((((...................))))..)))(((....))).))))).))))).)).))))))))........ (-26.98 = -27.40 + 0.42)
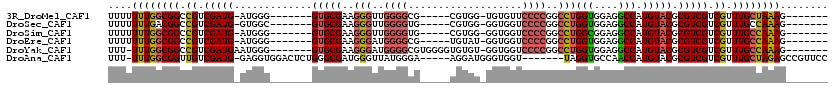
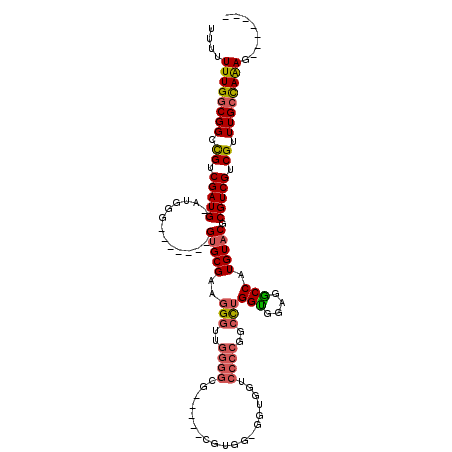
| Location | 7,157,824 – 7,157,923 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -17.42 |
| Energy contribution | -18.56 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7157824 99 - 27905053 -------CUUUAGCAAACGACGACGCGUACAUGGCCUCCACCAGGCCGGGGAACACA-CCACG-----CGCCCCAACCCUUCGCAC-------CCCAU-CAUCGACGGCCGCCAAAAAAA -------.....((...(((.((.(((.....(((((.....)))))((((..(...-....)-----..)))).......)))..-------....)-).)))...))........... ( -22.00) >DroSec_CAF1 90810 99 - 1 -------CUUUGGCAAACGACGACGCGUACAUGGCCUCCACCAGGCCGGGGACCACC-CCACG-----CACCCCAACCCUUCGCAC-------GCCAC-CAUCGACGGCCGUCAAAAAAA -------...........((((..((((....(((((.....)))))((((....))-)))))-----).................-------(((..-.......)))))))....... ( -30.30) >DroSim_CAF1 103984 99 - 1 -------CUUUGGCAAACGACGACGCGUACAUGGCCUCCGCCAGGCCGGGGACCACC-CCACG-----CACCCCAACCCUUCGCAC-------CCCAU-CAUCGACGGCCGCCAAAAAAA -------.((((((..........((((....(((((.....)))))((((....))-)))))-----)........((.(((...-------.....-...))).))..)))))).... ( -31.60) >DroEre_CAF1 92524 99 - 1 -------CUUUGGCAAACGACGACGCGUACAUGGCCUCCACCAGGCCGGGGACCACC-AUACA-----CGCCCCAUCCCUUCGCAC-------CCCAU-CAUCGACGGCCGCCAAAAAAA -------.((((((...((.(((.(((.....(((((.....)))))((((......-.....-----..)))).......)))..-------.....-..))).))...)))))).... ( -26.92) >DroYak_CAF1 94596 104 - 1 -------CUUUGGCAAACGACGACGCGUACAUGGCCUCCACCAGGCCGGGGACCACC-ACACACCCCACGCCCCAUCCCUUCGCAC-------CCCAUUCAUCGACGGCCGCCAAA-AAA -------.((((((..........((((....(((((.....)))))((((......-.....))))))))......((.(((...-------.........))).))..))))))-... ( -29.60) >DroAna_CAF1 99768 106 - 1 GGAACGGCUCUAGCAAACGACGACGCGUACAUGGUUGGCACCUA-------ACCACCCAUCCU-----UCCCAUAACCCAUCGCCCAGAGUCCACCUC-CAUCGACAACCGCCAAA-AAA (((..((((((.((..(((......)))...(((((((...)))-------))))........-----..............))..))))))....))-)................-... ( -19.80) >consensus _______CUUUGGCAAACGACGACGCGUACAUGGCCUCCACCAGGCCGGGGACCACC_ACACG_____CGCCCCAACCCUUCGCAC_______CCCAU_CAUCGACGGCCGCCAAAAAAA ........((((((...((.(((.........(((((.....)))))((((...................))))...........................))).))...)))))).... (-17.42 = -18.56 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:45 2006