
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,153,220 – 7,153,328 |
| Length | 108 |
| Max. P | 0.710664 |

| Location | 7,153,220 – 7,153,328 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 71.69 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
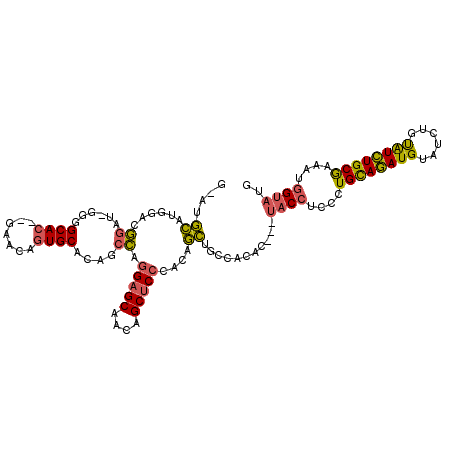
>3R_DroMel_CAF1 7153220 108 + 27905053 G-AUGCAUGGACGGAU-GGGGCACAUGAACAGUGCACAGCCAGGAGCAACAGCUCCCACAGCUGCCACAC---UACCUCCCUGCAGAUGUAUCUGUAUCUGCGAAAUGGUAUG .-((((..(((.((.(-((.((((.......)))).((((..(((((....)))))....)))).....)---))))))).((((((((......)))))))).....)))). ( -35.60) >DroVir_CAF1 134121 111 + 1 GAAUUGAGCGCGAAAUGGCGGCAG--CAACAAUGCAAAGCUAGGAGCAACAGCGCGCCCCCAAACCACACAUGUGCAUGUAUGUACAUGUGUGUGCGUGUGCAAAGUGGAUGG ...(((.(((((...(((.(((.(--((....)))...(((.(......))))..))).)))...((((((((((((....))))))))))))..))))).)))......... ( -39.70) >DroSec_CAF1 86133 106 + 1 G-AUGCAUGGACGGAU-GGGGCAC--GAACAGUGCACAGCCAGGAGCAACAGCUCCCACAGCUGCCACAC---UACCUCCCUGCAGAUGUAUCUGUAUCUGCGAUAUGGUAUG .-((((..(((.((.(-((.((((--.....)))).((((..(((((....)))))....)))).....)---))))))).((((((((......)))))))).....)))). ( -36.60) >consensus G_AUGCAUGGACGGAU_GGGGCAC__GAACAGUGCACAGCCAGGAGCAACAGCUCCCACAGCUGCCACAC___UACCUCCCUGCAGAUGUAUCUGUAUCUGCGAAAUGGUAUG ....((......((......((((.......))))....)).(((((....)))))....))...........((((....((((((((......))))))))....)))).. (-20.39 = -20.07 + -0.32)


| Location | 7,153,220 – 7,153,328 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 71.69 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -18.65 |
| Energy contribution | -21.10 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
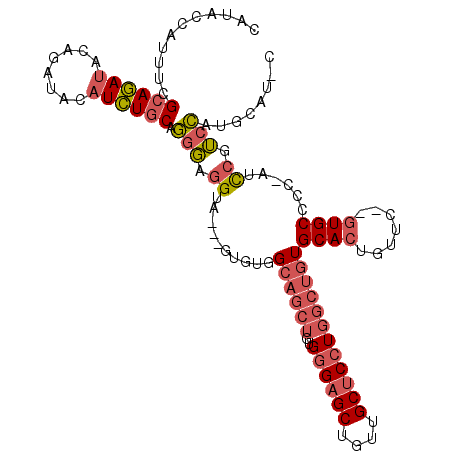
>3R_DroMel_CAF1 7153220 108 - 27905053 CAUACCAUUUCGCAGAUACAGAUACAUCUGCAGGGAGGUA---GUGUGGCAGCUGUGGGAGCUGUUGCUCCUGGCUGUGCACUGUUCAUGUGCCCC-AUCCGUCCAUGCAU-C ..((((.(..(((((((........)))))).)..)))))---(((((((((((..((((((....))))))))))))((((.......))))...-.......)))))..-. ( -41.50) >DroVir_CAF1 134121 111 - 1 CCAUCCACUUUGCACACGCACACACAUGUACAUACAUGCACAUGUGUGGUUUGGGGGCGCGCUGUUGCUCCUAGCUUUGCAUUGUUG--CUGCCGCCAUUUCGCGCUCAAUUC (((..(....(((....)))(((((((((.((....)).))))))))))..)))(((((((..((.((.....((...(((....))--).)).)).))..)))))))..... ( -37.80) >DroSec_CAF1 86133 106 - 1 CAUACCAUAUCGCAGAUACAGAUACAUCUGCAGGGAGGUA---GUGUGGCAGCUGUGGGAGCUGUUGCUCCUGGCUGUGCACUGUUC--GUGCCCC-AUCCGUCCAUGCAU-C ..((((...((((((((........)))))).))..))))---(((((((((((..((((((....))))))))))))((((.....--))))...-.......)))))..-. ( -39.00) >consensus CAUACCAUUUCGCAGAUACAGAUACAUCUGCAGGGAGGUA___GUGUGGCAGCUGUGGGAGCUGUUGCUCCUGGCUGUGCACUGUUC__GUGCCCC_AUCCGUCCAUGCAU_C ...........((((((........)))))).(((.((..........((((((..((((((....))))))))))))((((.......))))......)).)))........ (-18.65 = -21.10 + 2.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:41 2006