| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,141,004 – 7,141,110 |
| Length | 106 |
| Max. P | 0.558860 |

| Location | 7,141,004 – 7,141,110 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
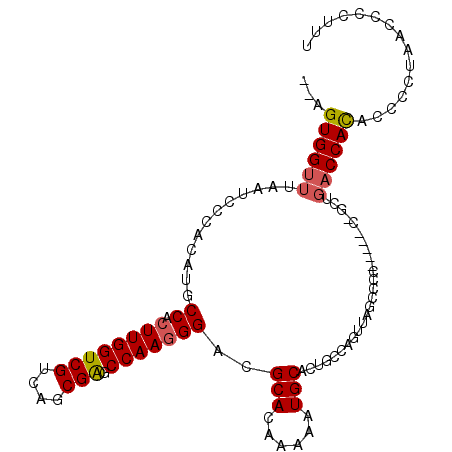
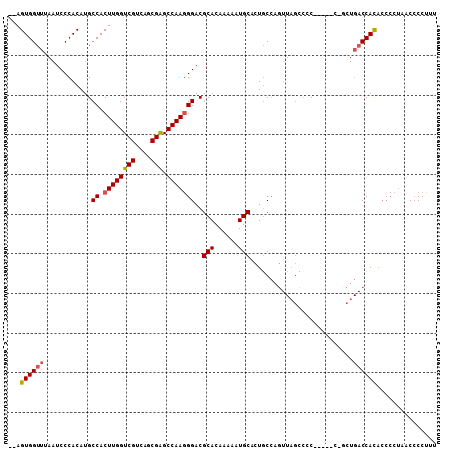
>3R_DroMel_CAF1 7141004 106 - 27905053 --AGUGGUUAAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGCCCCCCUACCCCCUGACCAUACCCCUUACCCCAUU --.(((((((...........((.((((((((....))).)))))))..(((......))).........................)))))))............... ( -22.10) >DroSec_CAF1 74041 105 - 1 --AGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGCCCCCCCU-CUGCUGACCACACCCCUCACCCCUUU --((((((............)))))).(((.(((((((((....(((..(((......)))...((......)))))..))-).))))))...)))............ ( -26.50) >DroSim_CAF1 84988 105 - 1 --AGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGUCCCCCCU-CCGCUGACCACACCCCUCACCCCUUU --((((((............)))))).(((.(((((((((....((((((((......)))((.....))..)))))..))-.)))))))...)))............ ( -30.00) >DroEre_CAF1 75270 97 - 1 --AGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGCCCC--------CUGACCACACCCCUACCACCCC- --.(((((.............((.((((((((....))).)))))))..(((......))).......(((((....--------))))).........)))))...- ( -22.80) >DroYak_CAF1 77281 97 - 1 --AGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGCC-C--------CUGACCACACCCCUAGCCCCCCU --.((((((.......((...((.((((((((....))).)))))))..(((......))).......)).....-.--------..))))))............... ( -22.86) >DroAna_CAF1 81636 84 - 1 CCAGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAACGGGCCAAAGGACGCACAA-AAUGCAAUCCU---------C-----CUGCU--CCAUC-------CCCCUGU ((((((((............))))).)))........((((...((((.(((...-..)))..))))---------.-----..)).--))...-------....... ( -18.70) >consensus __AGUGGUUUAAUCCCACAUGCCACUUGGUCGUCAGCGAGCCAAGGGACGCACAAAAAUGCACUGCCAGUUAGCCCC_____C_GCUGACCACACCCCUAACCCCUUU ...((((((............((.((((((((....))).)))))))..(((......)))..........................))))))............... (-20.14 = -20.28 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:31 2006