
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,136,166 – 7,136,258 |
| Length | 92 |
| Max. P | 0.528225 |

| Location | 7,136,166 – 7,136,258 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -15.32 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7136166 92 - 27905053 --C--G----AACUCCGGCU--------------------CCAACUUGUGUGUGUUCAUUAUGUGAACAUGCAGCUUAUUUGAUUUCUGCAUGCAAACUAGUCGACUAAGGACAUUUGCU --.--.----......(((.--------------------........(((((((((((...))))))))))).((((.((((((..((....))....)))))).)))).......))) ( -19.40) >DroVir_CAF1 93548 91 - 1 GAC--G----GCCUCAGAGU--------------------CCAGU---UUUAUGUUCAUUAUGUGUACAUGGAGCUUAUUUGAUUUCUGCAUGUAAAUUAGUCGACUGAGGCCAUUUGUU ...--(----(((((((...--------------------..(((---(((((((.(((...))).))))))))))...((((((..(........)..))))))))))))))....... ( -28.70) >DroSec_CAF1 68914 89 - 1 --C--G----AACUCCGGCU--------------------CCAACUUGUGUGUGUUCAUUAUGUGAACAUGCAGCUUAUUUGAUUUCUGCAUGCAAAUUAGUCG---AAGGACAUUUGCU --.--(----((.((.(...--------------------.)......(((((((((((...)))))))))))........)).))).....((((((..(((.---...))))))))). ( -21.50) >DroSim_CAF1 77239 92 - 1 --C--G----AACUCCGGCU--------------------CCAACUAGUGUGUGUUCAUUAUGUGAACAUGCAGCUUAUUUGAUUUCUGCAUGCAAAUUAGUCGACUAAGGACAUUUGCU --(--(----((.(((....--------------------...((((((.(((((((((...))))))((((((............))))))))).)))))).......)))..)))).. ( -19.44) >DroMoj_CAF1 85421 104 - 1 GGUCCGGGAGCUCGCUGAGC--------------UCGCUGCCAGU-U-UUUAUGUUCAUUAUGUGAACAUGGAGCUUAUUUGAUUUCUGCAUGUAAAUUACUCGACUGAGGCUAUUUGUU (((((((((((((((((.((--------------.....))))))-.-..(((((((((...)))))))))))))))...(((((((.....).)))))).....))).))))....... ( -29.30) >DroAna_CAF1 76633 112 - 1 --C--A----ACUUCUGGCUUCCAACUUCCCAACCAACUACCGACUUGUGUGUGUUCAUUAUGUGAACAUGCAGCUUAUUUGAUUUCUGCAUGCAAAUUAGUCGACUAAGGACAUUUGUU --.--.----.....(((...))).........((......(((((....(((((((((...))))))((((((............)))))))))....))))).....))......... ( -19.10) >consensus __C__G____AACUCCGGCU____________________CCAACUUGUGUGUGUUCAUUAUGUGAACAUGCAGCUUAUUUGAUUUCUGCAUGCAAAUUAGUCGACUAAGGACAUUUGCU ................................................(((((((((((...))))))))))).((((.((((((..((....))....)))))).)))).......... (-15.32 = -14.88 + -0.44)

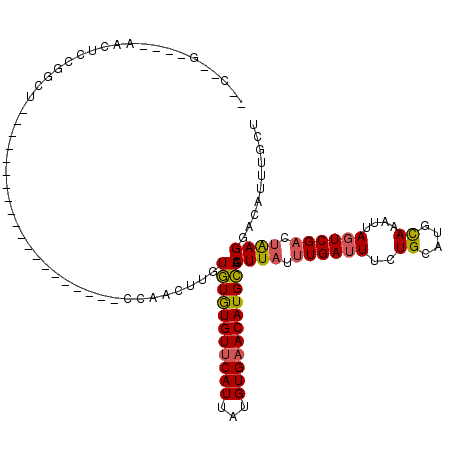
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:26 2006