| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,134,117 – 7,134,219 |
| Length | 102 |
| Max. P | 0.852776 |

| Location | 7,134,117 – 7,134,219 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.25 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -13.02 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.852776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
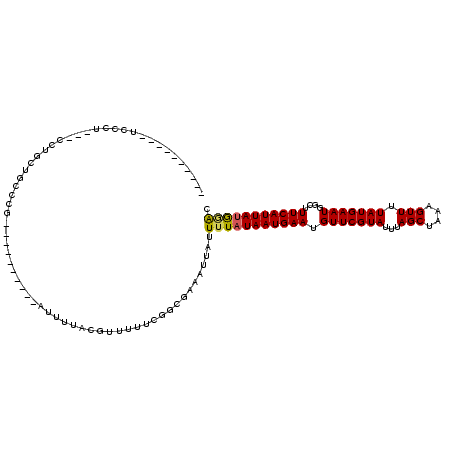
>3R_DroMel_CAF1 7134117 102 + 27905053 --------ACUCCCUUAUCCUGCUGCCCG----------AUUUCACGUAUUUCGGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCUUUCAUUAUGGAC --------....................(----------(((((.((.....))..))))))..(((((((((((.(((((((...(((....))).)))))))....))))))))))). ( -18.90) >DroVir_CAF1 91079 100 + 1 --------------C---CCCGUUGCGC---UCGCACCUAUCGUUCUUUUUAGGGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCGUUCAUUAUGGGC --------------.---(((.......---((((.((((..........)))))))).........((((((((((((((((...(((....))).)))))...)))))))))))))). ( -28.10) >DroGri_CAF1 80566 108 + 1 ---------CCCCCU---CCCGCUGCGCACCUUGCACUAUUCGUUCUUUGCGGAGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCGUUCAUUAUGGGC ---------..(((.---..((((.((((....((.......))....)))).))))..........((((((((((((((((...(((....))).)))))...)))))))))))))). ( -31.50) >DroEre_CAF1 68539 100 + 1 ----------UCCCAAAUCCUGGUGCCCG----------AUUUCACGUAUUUCGGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCUUUCAUUAUGCAC ----------............((((..(----------(((((.((.....))..)))))).....((((((((.(((((((...(((....))).)))))))....)))))))))))) ( -20.20) >DroWil_CAF1 71621 106 + 1 UGUGUAUGCCUCCCUCUUUCUGUAAC--------------UUUUACAUUUUUCGGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCUUUCAUUAUAAAA ......((((..........((((..--------------...))))......))))......((((((((((((.(((((((...(((....))).)))))))....)))))))))))) ( -18.19) >DroAna_CAF1 74656 86 + 1 ------------------------CCCUG----------AUUUUACGUUUUUCGCCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCCAAAGUUUUAUGAAUGGCUUUCAUUACGGGA ------------------------(((((----------(((((.((.....))..))))))).....(((((((.(((((((...(((....))).)))))))....))))))).))). ( -16.70) >consensus __________UCCCU___CCUGCUGCCCG__________AUUUUACGUUUUUCGGCGAAAUUAUUUUAUAAUGAAUGUUCGUAUUUAGCUAAAGUUUUAUGAAUGGCUUUCAUUAUGGAC ................................................................(((((((((((.(((((((...(((....))).)))))))....))))))))))). (-13.02 = -12.85 + -0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:23 2006