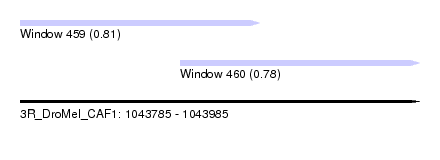
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,043,785 – 1,043,985 |
| Length | 200 |
| Max. P | 0.806556 |

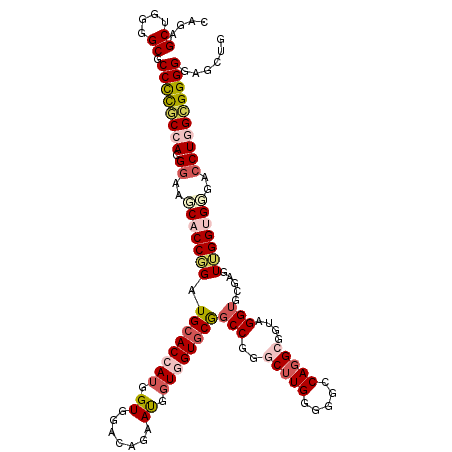
| Location | 1,043,785 – 1,043,905 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -56.34 |
| Consensus MFE | -31.56 |
| Energy contribution | -34.80 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1043785 120 + 27905053 CUGCGCGGGGCCUCCUGCCCUCUGGCGGCACCACUAGUGCCUGGUCCUCGCCCUGGCUCCUGGCUACCUCCUGGCUCGGGCAGAGCUAGGGCGCCCCGCCACGAAAGCACCGGAUGCACC .((((((((((..((.(((....)))(((((.....))))).)).....(((((((((((((((((.....)))).))))...))))))))))))))))..(....)........))).. ( -58.40) >DroSec_CAF1 15600 120 + 1 CUGCGCCGUACCUCCUGCCCUCUGGAGGCACCACUAGUGCCUGGCCCUGGCCCUGGCUCCUGCCUGCCUCCUGGCUCGCACAGAGCUGAGGCGCCCCGCCACGGAAGCACCGGAUGCACC .(((((((..(((((........)))))........((((....((.((((...(((....))).(((((..(((((.....)))))))))).....)))).))..))))))).)))).. ( -50.90) >DroSim_CAF1 15596 120 + 1 CUGCGCCGUACCUCCUGCCCUCUGGAGGCACCACUAGUGCCUGGCCCUGGCCCUGGCUCCUGGCUGCCUCCUGGCUCGCACAGAGCUGAGGCGCCCCGCCACGGAAGCACCGGAUGCACC .(((((((........((((((.((((((((((...(.(((.(((....)))..))).).))).))))))).(((((.....))))))))).)).(((...)))......))).)))).. ( -52.10) >DroEre_CAF1 15680 114 + 1 CUGCGCCGGGCCUCCUCCUCUCUGGAGGCUCUGCUACUCCCUG------GCACCGGACCCUGGCUGCCUGCUGGCGCGGGCCCAGCCGGGGCGCCUCUCCAGGGAACCACCUGGUGCACG .((((((((((((((........))))))))......((((((------(....((.((((((((((((((....))))))..))))))))..))...))))))).......)))))).. ( -66.80) >DroYak_CAF1 15405 114 + 1 CUGCGCCGGGCCUCCUCCUCUCUGGAGGCUCUGCCACUCCCUG------GCACUGGACACUGACUGCCUCCUGGCUUAGGCCCAGCUGGGGCGCCUUGCCACGGAACCACCUGGUGCACG .((((((((((((((........))))))))......(((.((------(((..((.(.((((..(((....)))))))((((.....))))))).))))).))).......)))))).. ( -53.50) >consensus CUGCGCCGGGCCUCCUGCCCUCUGGAGGCACCACUAGUGCCUGG_CCU_GCCCUGGCUCCUGGCUGCCUCCUGGCUCGGGCAGAGCUGGGGCGCCCCGCCACGGAAGCACCGGAUGCACC .(((...((((((((........)))))))).........((((......((.((((....(((.(((((..(((((.....)))))))))))))..)))).)).....))))..))).. (-31.56 = -34.80 + 3.24)

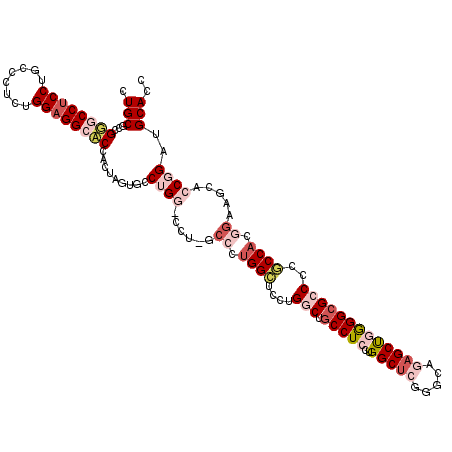
| Location | 1,043,865 – 1,043,985 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -55.84 |
| Consensus MFE | -37.50 |
| Energy contribution | -39.38 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1043865 120 + 27905053 CAGAGCUAGGGCGCCCCGCCACGAAAGCACCGGAUGCACCAAGGUGGAUAGAAUGGUGGUUCGGCCGGGCUUGGGUGACAGGCGAUAGGUGCGAGUUGGUGGGACCUGGCGGGGGAGCUG .........(((.(((((((((....)((((((.((((((..(((.((............)).)))..(((((.....)))))....))))))..)))))).....))))))))..))). ( -52.20) >DroSec_CAF1 15680 120 + 1 CAGAGCUGAGGCGCCCCGCCACGGAAGCACCGGAUGCACCAUGGUGGACAGAAUGGUGGUGCGUCCGGCCUUGGGGGCCAGGAGGUAGGUGUGAGUUGGUGUGACCUGGCGGGGGAGUUG ....((....)).((((((((.((..((((((((((((((((.((.......)).)))))))))))((((.....))))..................)))))..))))))))))...... ( -62.30) >DroSim_CAF1 15676 120 + 1 CAGAGCUGAGGCGCCCCGCCACGGAAGCACCGGAUGCACCAUGGUGGACAGAAUGGUGGUGCGGCCGACCUUGGGGGCCAGGCGGUAGGUGUGAGUUGGUGUGACCUGGCGGGGGAGUUG ....((....)).((((((((.((..(((((((.((((((((.((.......)).))))))))((((.(((.((...))))))))).........)))))))..))))))))))...... ( -56.00) >DroEre_CAF1 15754 120 + 1 CCCAGCCGGGGCGCCUCUCCAGGGAACCACCUGGUGCACGAUGGUGGACAGAACGGUGGUGCAGCCCGGCUUGAUAGCCAGGCGGUAGGUGCGACUCGGGGGGUCCUUGAGGGGGUGCUG (((....)))(((((((..((((((.((.((((((((((.((.((.......)).)).)))))(((.((((....)))).)))............))))))).))))))..))))))).. ( -60.50) >DroYak_CAF1 15479 120 + 1 CCCAGCUGGGGCGCCUUGCCACGGAACCACCUGGUGCACGAUAGUGGACAGGACGGUGGUGCAGCCCGGCUUGAUUGUCAUGCGGUUGGUCCGACUAGGGGAGUCCUUGAGGGGGAAGUG (((..(.(((((.((((....((((((((((..((.(((....))).)).....)))))).(((((..((.((.....)).))))))).))))....)))).))))).)..)))...... ( -48.20) >consensus CAGAGCUGGGGCGCCCCGCCACGGAAGCACCGGAUGCACCAUGGUGGACAGAAUGGUGGUGCGGCCGGGCUUGGGGGCCAGGCGGUAGGUGCGAGUUGGUGGGACCUGGCGGGGGAGCUG ....((....)).((((((((.((..(((((((.((((((((.((.......)).))))))))(((..(((((.....)))))....))).....)))))))..))))))))))...... (-37.50 = -39.38 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:01 2006