
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,129,506 – 7,129,661 |
| Length | 155 |
| Max. P | 0.996625 |

| Location | 7,129,506 – 7,129,600 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129506 94 + 27905053 GAUGCCGC-CUAUGUGUGUUCGUGUGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG ...((((.-..((((...((((((..(....)..))))))...)))).)))).(((((........)))))...(((((((((...))))))))) ( -28.40) >DroGri_CAF1 76084 80 + 1 G-----------A----GUGUGUGUGUGUGUUGACAUGCAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG (-----------(----((((.(((((((....)))))))...))))))....(((((........)))))...(((((((((...))))))))) ( -27.40) >DroSec_CAF1 62441 82 + 1 GCAGCCGC-CUGUG----------UGU--GUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG ...(((((-..(((----------(((--((((((((((....(((((.....)))))......))))))...))))))))))...))).))... ( -26.30) >DroSim_CAF1 69991 84 + 1 GCUGCCGC-CUGUG----------UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG ...(((((-..(((----------((((((((.((((((....(((((.....)))))......)))))).).))))))))))...))).))... ( -28.40) >DroEre_CAF1 63850 85 + 1 GCUGCCAUGCUAUG----------UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG ...((((((..(((----------((((((((.((((((....(((((.....)))))......)))))).).))))))))))..)))).))... ( -28.10) >DroAna_CAF1 70457 71 + 1 GCC------------------------GUGUGUGUGUGGAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG (((------------------------(.((((((.....)).)))).)))).(((((........)))))...(((((((((...))))))))) ( -22.90) >consensus GCUGCCGC_CU_UG__________UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUG ..............................((..(((((((.(((((..((..(((((........)))))))..)))))..)))))))..)).. (-17.48 = -17.20 + -0.28)



| Location | 7,129,506 – 7,129,600 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129506 94 - 27905053 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACACACGAACACACAUAG-GCGGCAUC ...(((((.....))))).....(.((((........)))))..(((.(((..((((((..............)))).)).)))..-.))).... ( -15.94) >DroGri_CAF1 76084 80 - 1 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUGCAUGUCAACACACACACACAC----U-----------C ...(((((.....))))).....((((((((((....(((.........))))))))))).))...............----.-----------. ( -16.70) >DroSec_CAF1 62441 82 - 1 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACAC--ACA----------CACAG-GCGGCUGC ...(((((.....))))).....(.((((........)))))........(((((..(((......--...----------.....-)))))))) ( -18.44) >DroSim_CAF1 69991 84 - 1 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA----------CACAG-GCGGCAGC ..(((........(((((.....(.((((........)))))....(((......))))))))......(.----------....)-)))..... ( -15.90) >DroEre_CAF1 63850 85 - 1 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA----------CAUAGCAUGGCAGC ...((........(((((.....(.((((........)))))....(((......))))))))........----------(((...)))))... ( -14.80) >DroAna_CAF1 70457 71 - 1 CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUCCACACACACAC------------------------GGC ...(((((.....))))).....(.((((........)))))..(((..((..((.....)).))..)------------------------)). ( -12.70) >consensus CACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA__________CA_AG_GCGGCAGC ...(((((.....))))).......(((.((((....(((.........))))))).)))................................... (-11.43 = -11.07 + -0.36)



| Location | 7,129,523 – 7,129,640 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -35.75 |
| Energy contribution | -36.70 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129523 117 + 27905053 UUCGUGUGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCU ((((((..(....)..))))))...(((((((((((((((........))......(((((((((...)))))))))......))))))))))))).....(((((.....))))). ( -42.50) >DroGri_CAF1 76087 112 + 1 UGUGUGUGUGUGUUGACAUGCAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCAAAAGCCAAUCGGGUGCCUGCUGCACA-----ACUGUU (((((((((((....))))))....(((((((((((((((........))......(((((((((...)))))))))......))))))))))))).....)))))-----...... ( -40.30) >DroSec_CAF1 62454 109 + 1 ------UGU--GUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCU ------...--(((.(..(((((((((....(((....)))...(((.((((((...((((((((...))))))))(((....)))....))))))..))))))))))))..)))). ( -41.50) >DroSim_CAF1 70004 111 + 1 ------UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCU ------.....(((.(..(((((((((....(((....)))...(((.((((((...((((((((...))))))))(((....)))....))))))..))))))))))))..)))). ( -41.50) >DroEre_CAF1 63864 111 + 1 ------UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCU ------.....(((.(..(((((((((....(((....)))...(((.((((((...((((((((...))))))))(((....)))....))))))..))))))))))))..)))). ( -41.50) >DroAna_CAF1 70460 108 + 1 ---------GUGUGUGUGUGGAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUUCUGCU ---------........(..((((.(((((((((((((((........))......(((((((((...)))))))))......)))))))))))))(((...)))....))))..). ( -37.70) >consensus ______UGUGUGUGUGUGCGAAACUGCAUUCGGUUGGUGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCU ...............(..(((((((((....(((....)))...(((.((((((...((((((((...))))))))(((....)))....))))))..))))))))))))..).... (-35.75 = -36.70 + 0.95)

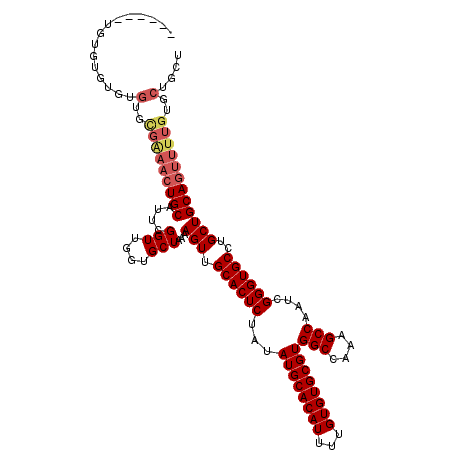

| Location | 7,129,523 – 7,129,640 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -27.68 |
| Energy contribution | -28.32 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129523 117 - 27905053 AGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACACACGAA ....((...((((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))).))................. ( -30.80) >DroGri_CAF1 76087 112 - 1 AACAGU-----UGUGCAGCAGGCACCCGAUUGGCUUUUGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUGCAUGUCAACACACACACACA ....((-----((((((((..(((..((..((((....)))).(((((.....))))).....(.((((........)))))...))..))).))))))...))))........... ( -31.90) >DroSec_CAF1 62454 109 - 1 AGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACAC--ACA------ ....((...((((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))).))......--...------ ( -30.80) >DroSim_CAF1 70004 111 - 1 AGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA------ ....((...((((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))).))...........------ ( -30.80) >DroEre_CAF1 63864 111 - 1 AGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA------ ....((...((((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))).))...........------ ( -30.80) >DroAna_CAF1 70460 108 - 1 AGCAGAACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUCCACACACACAC--------- ..........(((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))............--------- ( -27.90) >consensus AGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCACCAACCGAAUGCAGUUUCGCACACACACACA______ ....((...((((((((...((........((((....)))).(((((.....))))).....(.((((........)))))..))...)))))))).))................. (-27.68 = -28.32 + 0.64)



| Location | 7,129,560 – 7,129,661 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129560 101 + 27905053 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCUUACUUUUUCUCAACC-----CCCCCC ((..((((((.((((((...((((((((...))))))))(((....)))....)))))).....(((((.....)))))..))))))...))...-----...... ( -29.80) >DroVir_CAF1 86881 92 + 1 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCAAAAGCCAAUCGGGUGCUUGCUGCACA-----ACUGCUUAUUUUUCAUCCG---------ACCC .......(((.((((((...((((((((...))))))))(((....)))....))))))..)))(((..-----..)))..............---------.... ( -25.10) >DroGri_CAF1 76124 92 + 1 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCAAAAGCCAAUCGGGUGCCUGCUGCACA-----ACUGUUUCUUUUUCCACAG---------UCCC (((....(((.((((((...((((((((...))))))))(((....)))....))))))..))))))..-----(((((..........))))---------)... ( -26.70) >DroSec_CAF1 62483 95 + 1 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCUUACUUUUUCUCAACC----------- ((..((((((.((((((...((((((((...))))))))(((....)))....)))))).....(((((.....)))))..))))))...))...----------- ( -29.80) >DroWil_CAF1 67694 105 + 1 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUACGCGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGCUCUGUUUACCAUUUUCAGAUCGCCUCGUG-CC .......(((.((((((....(((((.....)))))...(((....)))....))))))..)))((((...((.((((...........)))).))...).))-). ( -24.00) >DroYak_CAF1 65718 106 + 1 UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCUUACUUUUUCUCAACCAAACACCCCAC ((..((((((.((((((...((((((((...))))))))(((....)))....)))))).....(((((.....)))))..))))))...)).............. ( -29.80) >consensus UGCUAAAAGUUGCACUCUAUAUGCACAUUUUGUGUGCGUGGCCAAAGCCAAUCGGGUGCCUGCUGCAGUUUUGUGCUGCUUACUUUUUCUCAACC_______CCCC .......(((.((((((...((((((((...))))))))(((....)))....))))))..)))((((.......))))........................... (-24.47 = -24.92 + 0.45)

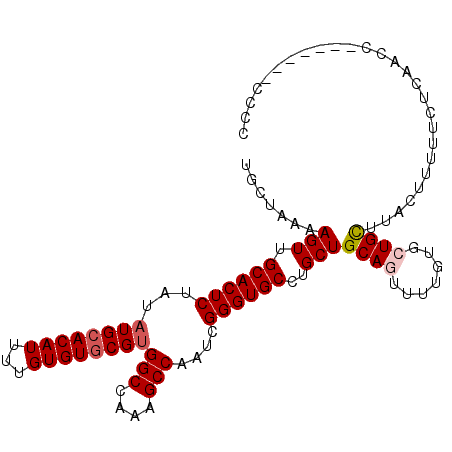

| Location | 7,129,560 – 7,129,661 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7129560 101 - 27905053 GGGGGG-----GGUUGAGAAAAAGUAAGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA ......-----.(((((((........((((.......)))).....((((.(...((((....)))).(((((.....))))).....).))))...))))))). ( -27.40) >DroVir_CAF1 86881 92 - 1 GGGU---------CGGAUGAAAAAUAAGCAGU-----UGUGCAGCAAGCACCCGAUUGGCUUUUGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA ....---------..............(((((-----((..(..............((((....)))).(((((.....))))).......)..)))))....)). ( -22.40) >DroGri_CAF1 76124 92 - 1 GGGA---------CUGUGGAAAAAGAAACAGU-----UGUGCAGCAGGCACCCGAUUGGCUUUUGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA .(.(---------((((((.(((((...((((-----(((((.....))))..))))).))))).))))(((((.....)))))......))).)........... ( -27.40) >DroSec_CAF1 62483 95 - 1 -----------GGUUGAGAAAAAGUAAGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA -----------.(((((((........((((.......)))).....((((.(...((((....)))).(((((.....))))).....).))))...))))))). ( -27.40) >DroWil_CAF1 67694 105 - 1 GG-CACGAGGCGAUCUGAAAAUGGUAAACAGAGCAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCGCGUACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA .(-((((.(((.(((.......)))...((((((.....(((.....)))........))))))))).)(((((.....))))).......))))........... ( -25.22) >DroYak_CAF1 65718 106 - 1 GUGGGGUGUUUGGUUGAGAAAAAGUAAGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA ((((((((((((.((......))....((((.......))))..)))))))))(..((((....))))..).)))((((..(((((.....)))))..)))).... ( -31.00) >consensus GGGG_______GGUUGAGAAAAAGUAAGCAGCACAAAACUGCAGCAGGCACCCGAUUGGCUUUGGCCACGCACACAAAAUGUGCAUAUAGAGUGCAACUUUUAGCA ...................((((((..((((.......)))).....((((.(...((((....)))).(((((.....))))).....).)))).)))))).... (-19.86 = -20.78 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:18 2006