
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,111,663 – 7,111,757 |
| Length | 94 |
| Max. P | 0.858606 |

| Location | 7,111,663 – 7,111,757 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
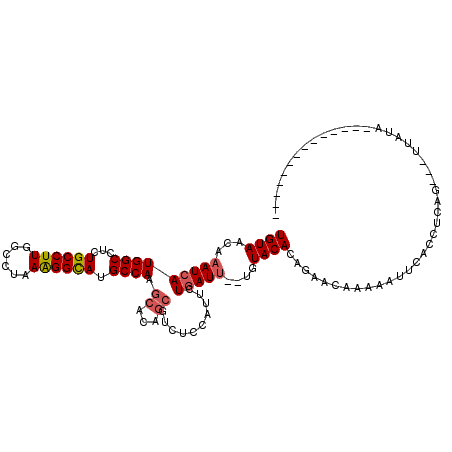
>3R_DroMel_CAF1 7111663 94 + 27905053 UGUAACAAAUCAUGGCCACUGCCUUGGCCUAAAGGCAUGCCAAGCACAGCGUCUCCAUUCUGAUU--UGUACACAGAGAGAAAAUUCACUU-AG---UUA---------------- .(((.(((((((((((...((((((......)))))).)))).((...))..........)))))--)))))((.(((.((....)).)))-.)---)..---------------- ( -23.20) >DroSec_CAF1 44673 111 + 1 UGUAACAAAUCAUGGCCUCUGCCUUGGCCUAAAGGCAUGCCAAGCACAGCGUCUCCGUUCUGAUU--CGUACACAGAACGAAAAUUCACCUCAG---UUAUAUUGCCGGUAGAGUU ............((((...((((((......)))))).)))).............(((((((...--......)))))))..((((((((.(((---(...))))..))).))))) ( -26.60) >DroSim_CAF1 50726 111 + 1 UGUAACAAAUCAUGGCCUCUGCCUUGGCCUAAAGGCAUGCCAAGCACAGCGUCUCCAUUCUGAUU--CGUACACAGAACGAAAAUUCACCUCAG---UUAUAUUGCUGGUAAAGUU ...(((......((((...((((((......)))))).)))).((.(((((........((((((--(((.......)))))........))))---......)))))))...))) ( -25.64) >DroEre_CAF1 45926 97 + 1 UGUAACAAAUCAUGGCCUCUGCCUUGGCCUAAAGGCAUGCCAAGCACAGCGUCUCCAUUCUGAUU--UUUACACUGACCAGAAAAAAAGCACUG---UUAUG-------------- .((((((.....((((...((((((......)))))).))))......((.......(((((.((--........)).))))).....))..))---)))).-------------- ( -22.80) >DroYak_CAF1 47120 85 + 1 UGUAACAAAUCAUGGCCUCUGCCUUGGCCCAAGGGCAUGCCAAGCACAGCGUCUCCAUUCUGAUU--UGUACACUGAUCAAAAAAGA----------------------------- ((((.(((((((.((((........))))...(((.((((........)))).)))....)))))--))))))..............----------------------------- ( -22.90) >DroAna_CAF1 51639 114 + 1 UGUAACAAAUCAUGGCAGCUGCCUUGGCCAAAAGGUAUGCCAUGGCAUGAAUCUC-AGUCUCAUCGCUUUACACAGGAAAAGUA-UCAUUCUAGAGUUUAAAAUCUUGACAAACUU (((((.....(((((((..((((((......)))))))))))))((((((.....-....)))).)).)))))(((((......-((......))........)))))........ ( -25.34) >consensus UGUAACAAAUCAUGGCCUCUGCCUUGGCCUAAAGGCAUGCCAAGCACAGCGUCUCCAUUCUGAUU__UGUACACAGAACAAAAAUUCACCUCAG___UUAUA______________ ((((...(((((((((...((((((......)))))).)))).((...))..........)))))....))))........................................... (-15.71 = -16.10 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:02 2006