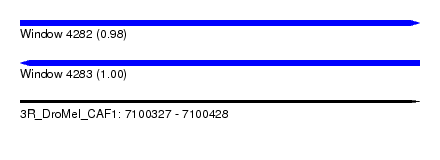
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,100,327 – 7,100,428 |
| Length | 101 |
| Max. P | 0.998607 |

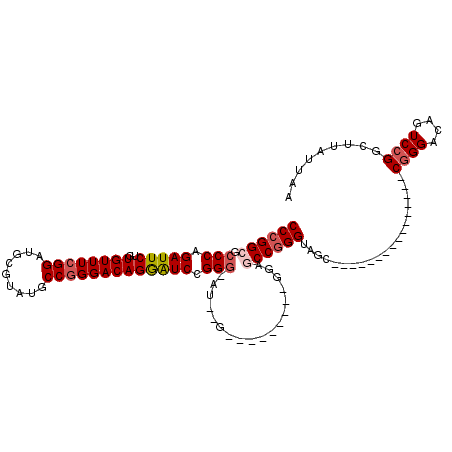
| Location | 7,100,327 – 7,100,428 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -44.46 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7100327 101 + 27905053 CCCGGCGCCCAGAUUCUGUGUUUCGGAUGCGUAUGCCGGGACAGGAUCCGGGUAUCUGGAGAUUCGUGGAGCCGGGUGGC----------------CGGGAGAGUCCGGCUUAUUAA ((((((..(((((.(((.(((..(((.........)))..))).).(((((....))))))).)).))).)))))).(((----------------((((....)))))))...... ( -44.90) >DroSec_CAF1 33345 85 + 1 CCCGGCGCCCAGAUUCUGUGUUUCGGAUGCGUAUGCCGGGACAGGAUCCGGG----------------GAGCCGGGUAGC----------------CGGGACAGUCCGGCUUAUUAA ((((((.(((.((((((((..(((((.........))))))))))))).)))----------------..)))))).(((----------------((((....)))))))...... ( -43.70) >DroSim_CAF1 37208 85 + 1 CCCGGCGCCCAGAUUCUGUGUUUCGGAUGCGUAUGCCCGGACAGGAUCCGGG----------------UGGCCGGGUAGC----------------CGGGACAGUCCGGCUUAUUAA ((((((((((.((((((((...((((..((....)))))))))))))).)))----------------).)))))).(((----------------((((....)))))))...... ( -44.90) >DroEre_CAF1 34133 109 + 1 CCCGGCGCCCAGAUUCUGUGUUUCGGAUGCGUAUGCCGGGACAGGAUCCGGGGAUCCG--------GGGACCCAGGGAUCCAGUGAAUCGGGGAGCCAGGACAGUCCGGCUUAUUAA .(((((.(((.(((((..(((..(((.........)))..)))((((((.(((.((..--------..)))))..))))))...))))).))).))).((.....))))........ ( -48.70) >DroYak_CAF1 35512 93 + 1 CCCGGCGCCCAGAUUCAGUGUUUCGGAUGCGUAUGCCGGGACAGAGUCCGGUGAUUCG--------GGGCUCCGGGAAUC----------------CGGGACAGUCCGGCUUAUUAA (((((.((((.(((((..(((..(((.........)))..))))))))(((....)))--------)))).)))))...(----------------((((....)))))........ ( -40.10) >consensus CCCGGCGCCCAGAUUCUGUGUUUCGGAUGCGUAUGCCGGGACAGGAUCCGGG_AU__G_________GGAGCCGGGUAGC________________CGGGACAGUCCGGCUUAUUAA ((((((.(((.(((((..((((((((.........))))))))))))).)))..................))))))....................((((....))))......... (-25.27 = -26.15 + 0.88)

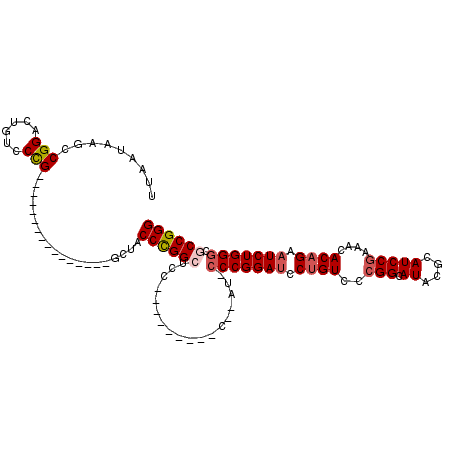
| Location | 7,100,327 – 7,100,428 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -21.51 |
| Energy contribution | -22.39 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7100327 101 - 27905053 UUAAUAAGCCGGACUCUCCCG----------------GCCACCCGGCUCCACGAAUCUCCAGAUACCCGGAUCCUGUCCCGGCAUACGCAUCCGAAACACAGAAUCUGGGCGCCGGG .......(((((......)))----------------))..((((((.......(((....))).(((((((.((((..(((.((....)))))....)))).))))))).)))))) ( -38.90) >DroSec_CAF1 33345 85 - 1 UUAAUAAGCCGGACUGUCCCG----------------GCUACCCGGCUC----------------CCCGGAUCCUGUCCCGGCAUACGCAUCCGAAACACAGAAUCUGGGCGCCGGG ......((((((......)))----------------))).((((((..----------------(((((((.((((..(((.((....)))))....)))).))))))).)))))) ( -40.70) >DroSim_CAF1 37208 85 - 1 UUAAUAAGCCGGACUGUCCCG----------------GCUACCCGGCCA----------------CCCGGAUCCUGUCCGGGCAUACGCAUCCGAAACACAGAAUCUGGGCGCCGGG ......((((((......)))----------------))).((((((..----------------(((((((.((((.(((((....))..)))....)))).))))))).)))))) ( -42.20) >DroEre_CAF1 34133 109 - 1 UUAAUAAGCCGGACUGUCCUGGCUCCCCGAUUCACUGGAUCCCUGGGUCCC--------CGGAUCCCCGGAUCCUGUCCCGGCAUACGCAUCCGAAACACAGAAUCUGGGCGCCGGG .................((((((.(((.(((((...((((((..(((((..--------..)))))..))))))(((..(((.((....)))))..)))..))))).))).)))))) ( -45.10) >DroYak_CAF1 35512 93 - 1 UUAAUAAGCCGGACUGUCCCG----------------GAUUCCCGGAGCCC--------CGAAUCACCGGACUCUGUCCCGGCAUACGCAUCCGAAACACUGAAUCUGGGCGCCGGG ........((((......)))----------------)...(((((.((((--------.((.(((((((........))))....((....))......))).)).)))).))))) ( -34.10) >consensus UUAAUAAGCCGGACUGUCCCG________________GCUACCCGGCUCC_________C__AU_CCCGGAUCCUGUCCCGGCAUACGCAUCCGAAACACAGAAUCUGGGCGCCGGG .........(((......)))....................((((((..................(((((((.((((..(((.((....)))))....)))).))))))).)))))) (-21.51 = -22.39 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:00 2006