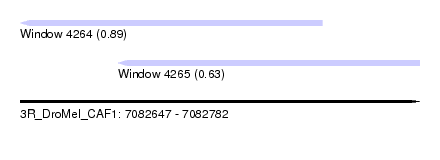
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,082,647 – 7,082,782 |
| Length | 135 |
| Max. P | 0.889098 |

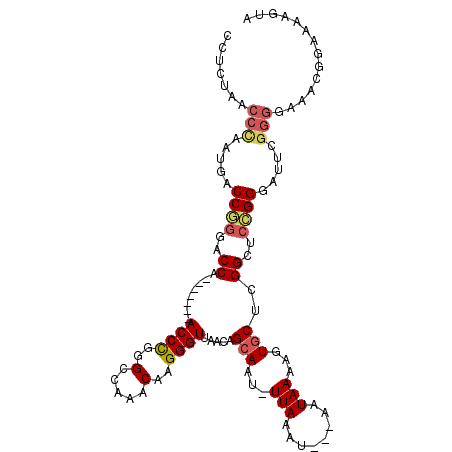
| Location | 7,082,647 – 7,082,749 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.15 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7082647 102 - 27905053 AAACAAGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAAAA--AAAACAGCGGCGCAACCCUUAACUUGU ....(((((((....((..((((.....)))).(((((((.(((((.....)))))...))....))))).....--......)).....)))))))....... ( -22.20) >DroSec_CAF1 15577 100 - 1 AAACAAGGGUUAACAGCAACUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAA----AAAACAGCGGCGCAACCCUUAACUUGU ....(((((((..(((((.(((.........))))))).)((.(((...((((......))))...((.....----...))..))).)))))))))....... ( -21.80) >DroSim_CAF1 18217 102 - 1 AAACAAGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAAAA--AAAACAGCGGCGCAACCCUUAACUUGU ....(((((((....((..((((.....)))).(((((((.(((((.....)))))...))....))))).....--......)).....)))))))....... ( -22.20) >DroEre_CAF1 15814 98 - 1 AAACAAGGGUUAAAAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAA------AAAAGCGGUGCAACCCUUAACUUGU ....(((((((.(..((..((((.....)))).(((((((.(((((.....)))))...))....)))))...------....))..)..)))))))....... ( -22.80) >DroYak_CAF1 16797 104 - 1 AAACAAGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAAAAAAAAAAAAGCGGCGCAACCCUUAACUUGU ....(((((((....((..((((.....)))).(((((((.(((((.....)))))...))....))))).............)).....)))))))....... ( -22.20) >DroAna_CAF1 17257 96 - 1 AAACAAGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCUGGGAUUCGGGGAAGCGAAACGG---AA-----AAAAAUUCAGCGCAACCCUUAACUUGU ....(((((((..(((((.(((........))).)))).).(((((.....)))))..(((.....(---((-----.....)))..))))))))))....... ( -19.60) >consensus AAACAAGGGUUAACAGCAAUUUAAAUAAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUACAAA____AAAAAAGCGGCGCAACCCUUAACUUGU ....(((((((....(((.(((........))).)))(((.(((((.....)))))...)))............................)))))))....... (-19.26 = -19.15 + -0.11)

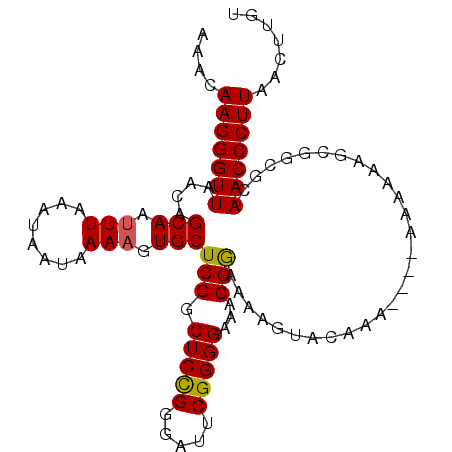
| Location | 7,082,680 – 7,082,782 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7082680 102 - 27905053 CCUCUAACCCAAUGACCGGGACCA-------ACCUGGGCCAAACAAGGGUUAACAGCAAU-UUAAAU---AAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUA ((.....((((((..((((..(((-------((((..(.....)..)))))...((((.(-((....---....))).)))).))..)))).))).))).....))....... ( -26.50) >DroVir_CAF1 29339 90 - 1 CCUCUAACCCGUUAC------CCA-------ACCCGGGCCAAGCAGGGGUUAACGGCAACUUUAAAUAAUAAUAAACGUACGUGGCAAC------C----AAACAAAAAAAAA (((((..((((....------...-------...))))...)).)))((((.(((...((((((........)))).)).)))...)))------)----............. ( -13.60) >DroSec_CAF1 15608 102 - 1 CCUCUAACCCAAUGACCGGGACCA-------ACCCGGGCCAAACAAGGGUUAACAGCAAC-UUAAAU---AAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUA ((.....((((((..((((..(((-------((((..(.....)..)))))...((((.(-((....---.....))))))).))..)))).))).))).....))....... ( -28.90) >DroEre_CAF1 15843 102 - 1 CCUCUAACCCAAUGACCGGGACCA-------ACCCGGGCCAAACAAGGGUUAAAAGCAAU-UUAAAU---AAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUA ((.....((((((..((((..(((-------((((..(.....)..)))))...((((.(-((....---....))).)))).))..)))).))).))).....))....... ( -28.40) >DroYak_CAF1 16832 102 - 1 CCUCUAACCCAAUGACCGGGACCA-------ACCCGGGCCAAACAAGGGUUAACAGCAAU-UUAAAU---AAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUA ((.....((((((..((((..(((-------((((..(.....)..)))))...((((.(-((....---....))).)))).))..)))).))).))).....))....... ( -28.40) >DroAna_CAF1 17286 106 - 1 CCUCUAACCU-AUGACCAGGACCAAGGACCUACCCAGGCCAAACAAGGGUUAACAGCAAU-UUAAAU---AAUAAAAGUGCUCGGCUCUGGGAUUCGGGGAAGCGAAACGG-- ((((......-......(((.(....).))).((((((((.(((....)))...((((.(-((....---....))).)))).))).)))))....))))...........-- ( -25.20) >consensus CCUCUAACCCAAUGACCGGGACCA_______ACCCGGGCCAAACAAGGGUUAACAGCAAU_UUAAAU___AAUAAAAGUGCUCGGCUCCGGGAUUCGGGGAAACGGAAAAGUA .......(((.....((((..((........((((..(.....)..)))).....(((...(((........)))...)))..))..)))).....))).............. (-18.27 = -18.88 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:42 2006