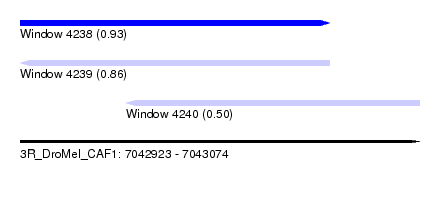
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,042,923 – 7,043,074 |
| Length | 151 |
| Max. P | 0.934236 |

| Location | 7,042,923 – 7,043,040 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -16.38 |
| Energy contribution | -15.38 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7042923 117 + 27905053 GAAGAAGCAAGAACACGGCCAAAAUAAAGUCCAAGAAGCACACAAUGCGCGUGGGCCGAAAGCUGAUCCAUGUAAAAGUCAUCGAUGAUAAGCAGCCAAAGCGUAUUGUGGACCGCA ......((........((.(........).))........((((((((((...(((.....(((.(((.(((.......))).)))....))).)))...))))))))))....)). ( -29.00) >DroPse_CAF1 439 116 + 1 GUC-UAACAAAACGAAGGGCAAAAUGAAGGCCAAUAAGCAAACAAUGCGUCUGGGACGAAAACUCAUACACGUCAAGGUGAUUGACGAUGCUAAUCCCAAACGCAUUGUGGAUCGGG ...-........(((..(((.........))).........(((((((((.(((((.((....)).....((((((.....)))))).......))))).)))))))))...))).. ( -34.40) >DroEre_CAF1 596 117 + 1 GAAGAAUCAAUAACUCGGCCAAAAUCAAAUCCAAGAAGCACACAAUGCGCAUGGGCCGAAAGCUAAUCCAUGUGAAAGUCAUCGAUGAUAAGCAACCAAAGCGUAUUGUGGACCGAA ..............((((.(....((........))....(((((((((((((((.(....)....))))).((...((((....))))...))......))))))))))).)))). ( -26.60) >DroYak_CAF1 602 117 + 1 GAAGAAUCAAGAACUCGGCCAAAAUCAAAUCCAAGAAGCACACAAUGCGCAUGGGCCGAAAGCUUAUUCAUGUGAAAGUCAUCGAUGAUAAGCAGCCAAAGCGCAUUGUGGACCGAA ..............((((.(....((........))....((((((((((...(((.....((((((.((((((.....)))..))))))))).)))...))))))))))).)))). ( -34.20) >DroAna_CAF1 374 111 + 1 ------GCAAGAUGAAGGUGAAGAAUAAGUCCAAUAAAAAAACGAUGCGCCUGGGCCGAAAGCUGAUUCACGUUAAAGUCAUUGACGACAAACAACCAAAGCGCAUUGUGGACCGGA ------......................((((.........(((((((((.((((.(....)).......((((((.....))))))........)))..))))))))))))).... ( -24.90) >DroPer_CAF1 613 116 + 1 GUC-AAACAAAACGAAGGGCAAAAUGAAGGCCAAUAAGCAAACAAUGCGCCUGGGACGAAAACUCAUACACGUCAAGGUGAUUGACGAUGCUAAUCCCAAACGCAUUGUGGAUCGGG ...-........(((..(((.........))).........((((((((..(((((.((....)).....((((((.....)))))).......)))))..))))))))...))).. ( -32.20) >consensus GAA_AAACAAGAACAAGGCCAAAAUCAAGUCCAAGAAGCAAACAAUGCGCCUGGGCCGAAAGCUCAUACACGUCAAAGUCAUCGACGAUAAGCAACCAAAGCGCAUUGUGGACCGGA .........................................(((((((((...((((((..(((............)))..)))..........)))...)))))))))........ (-16.38 = -15.38 + -1.00)



| Location | 7,042,923 – 7,043,040 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.37 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -15.86 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7042923 117 - 27905053 UGCGGUCCACAAUACGCUUUGGCUGCUUAUCAUCGAUGACUUUUACAUGGAUCAGCUUUCGGCCCACGCGCAUUGUGUGCUUCUUGGACUUUAUUUUGGCCGUGUUCUUGCUUCUUC .((((((((((((.(((...(((((......(((.(((.......))).))).......)))))...))).))))))..((....))..........)))))).............. ( -28.52) >DroPse_CAF1 439 116 - 1 CCCGAUCCACAAUGCGUUUGGGAUUAGCAUCGUCAAUCACCUUGACGUGUAUGAGUUUUCGUCCCAGACGCAUUGUUUGCUUAUUGGCCUUCAUUUUGCCCUUCGUUUUGUUA-GAC ..(((...((((((((((((((((..(((.((((((.....)))))))))..(......))))))))))))))))).........(((.........)))..)))........-... ( -35.90) >DroEre_CAF1 596 117 - 1 UUCGGUCCACAAUACGCUUUGGUUGCUUAUCAUCGAUGACUUUCACAUGGAUUAGCUUUCGGCCCAUGCGCAUUGUGUGCUUCUUGGAUUUGAUUUUGGCCGAGUUAUUGAUUCUUC (((((((((((((.(((...(((((......(((.(((.......))).))).......)))))...))).))))))....((........))....)))))))............. ( -27.42) >DroYak_CAF1 602 117 - 1 UUCGGUCCACAAUGCGCUUUGGCUGCUUAUCAUCGAUGACUUUCACAUGAAUAAGCUUUCGGCCCAUGCGCAUUGUGUGCUUCUUGGAUUUGAUUUUGGCCGAGUUCUUGAUUCUUC ...((..((((((((((...(((((((((((((.(.((.....)))))).))))))....))))...))))))))))..))((..(((((((........)))))))..))...... ( -39.40) >DroAna_CAF1 374 111 - 1 UCCGGUCCACAAUGCGCUUUGGUUGUUUGUCGUCAAUGACUUUAACGUGAAUCAGCUUUCGGCCCAGGCGCAUCGUUUUUUUAUUGGACUUAUUCUUCACCUUCAUCUUGC------ ...(((((((.((((((((.(((((((.((((....))))...)))..(((......))))))).)))))))).)).........))))).....................------ ( -26.10) >DroPer_CAF1 613 116 - 1 CCCGAUCCACAAUGCGUUUGGGAUUAGCAUCGUCAAUCACCUUGACGUGUAUGAGUUUUCGUCCCAGGCGCAUUGUUUGCUUAUUGGCCUUCAUUUUGCCCUUCGUUUUGUUU-GAC ..(((...((((((((((((((((..(((.((((((.....)))))))))..(......))))))))))))))))).........(((.........)))..)))........-... ( -35.40) >consensus UCCGGUCCACAAUGCGCUUUGGAUGCUUAUCAUCAAUGACUUUCACAUGAAUCAGCUUUCGGCCCAGGCGCAUUGUGUGCUUAUUGGACUUCAUUUUGGCCGUCUUCUUGAUU_UUC ...((((((((((((((...(((((..........(((.......)))...........)))))...))))))))).........)))))........................... (-15.86 = -16.42 + 0.56)

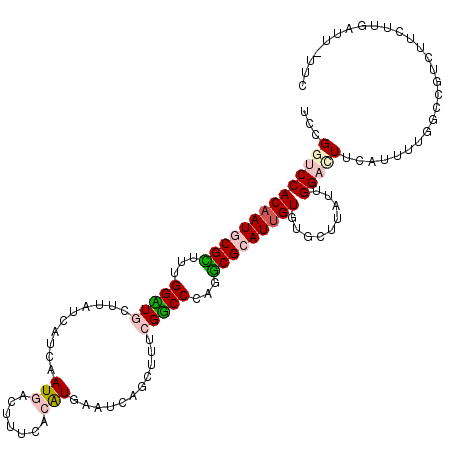
| Location | 7,042,963 – 7,043,074 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.62 |
| Mean single sequence MFE | -37.36 |
| Consensus MFE | -19.77 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7042963 111 - 27905053 UGCUCGCAGAUGCAGGGCUUGGCGCUGGGUCCAUUGCGGUCCACAAUACGCUUUGGCUG---CUUAUCAUCGAUGACUUUUACAUGGAUCAGCUUUCGGCCCACGCGCAUUGUG .((((((....)).))))...((((((((((((..(((..........)))..))((((---......(((.(((.......))).)))))))....)))))).))))...... ( -36.90) >DroPse_CAF1 478 111 - 1 UGCUCACAGAUACAGGGCUUGGCGAUUUGGCUGUCCCGAUCCACAAUGCGUUUGGGAUU---AGCAUCGUCAAUCACCUUGACGUGUAUGAGUUUUCGUCCCAGACGCAUUGUU ........(((...(((..((((......)))).))).))).((((((((((((((((.---.(((.((((((.....)))))))))..(......))))))))))))))))). ( -41.80) >DroWil_CAF1 343 114 - 1 UGCUCACAUAUGCAGGGUUUGGCCGUUUGACCAUCCCGAUCCAUAAUCCGUUUGGGUUGCUCCUUAUCAUCAAUUACUUUCACAUGGAUGAGUUUGCGUCCCAGACGCAUGGUU (((........)))(((..(((........))).)))...((((....((((((((.(((..((((((..................))))))...))).)))))))).)))).. ( -32.27) >DroYak_CAF1 642 111 - 1 UGCUCGCAGAUGCAGGGCUUGGCACUGGGUCCAUUUCGGUCCACAAUGCGCUUUGGCUG---CUUAUCAUCGAUGACUUUCACAUGAAUAAGCUUUCGGCCCAUGCGCAUUGUG .((((((....)).))))..((.((((((.....))))))))(((((((((...(((((---((((((((.(.((.....)))))).))))))....))))...))))))))). ( -40.60) >DroAna_CAF1 408 111 - 1 UGUUCGCAGAUGCAGGGCUUCGCUGUGGUGCUAUUCCGGUCCACAAUGCGCUUUGGUUG---UUUGUCGUCAAUGACUUUAACGUGAAUCAGCUUUCGGCCCAGGCGCAUCGUU .((((((....)).)))).....(((((.(((.....))))))))((((((((.(((((---((.((((....))))...)))..(((......))))))).)))))))).... ( -31.30) >DroPer_CAF1 652 111 - 1 UGCUCACAGAUACAGGGCUUGGCGAUUUGGCUGUCCCGAUCCACAAUGCGUUUGGGAUU---AGCAUCGUCAAUCACCUUGACGUGUAUGAGUUUUCGUCCCAGGCGCAUUGUU ........(((...(((..((((......)))).))).))).((((((((((((((((.---.(((.((((((.....)))))))))..(......))))))))))))))))). ( -41.30) >consensus UGCUCACAGAUGCAGGGCUUGGCGAUGGGGCCAUCCCGAUCCACAAUGCGCUUGGGAUG___CUUAUCAUCAAUGACUUUCACAUGAAUGAGCUUUCGGCCCAGGCGCAUUGUU ..............((((((((........)))....)))))(((((((((((.(((((...((((((((.............))).)))))....))))).))))))))))). (-19.77 = -20.30 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:18 2006