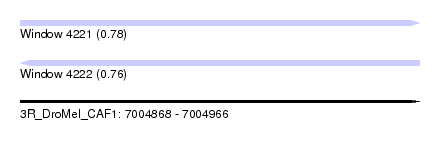
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 7,004,868 – 7,004,966 |
| Length | 98 |
| Max. P | 0.777687 |

| Location | 7,004,868 – 7,004,966 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -33.03 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
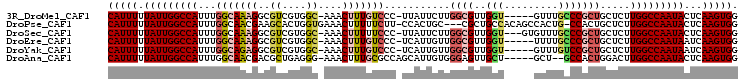
>3R_DroMel_CAF1 7004868 98 + 27905053 CAUUUUUAUUGGCCAUUUGGCAAAGGCGUCGUGGC-AAACUUUGUCCC-UUAUUCUUGGCGUUGGU-----GUUUGCCCGCUGCUCUUGGCCAAUACUCAAGUGG (((((.(((((((((...((((...(((....(((-((((.....(((-........)).).....-----))))))))))))))..)))))))))...))))). ( -33.70) >DroPse_CAF1 4945 100 + 1 CAUUUUUAUUGGCCAUUUGGCAACGAAGCACUGGUGAAACUUUUUCUU-CCACUGC---CGCUGCCACAGCCACUG-CCACUGCUCUUGGCCAAUACUCAAGUGG (((((.(((((((((..(((((.....(((.(((.(((.....)))..-))).)))---.((((...))))...))-))).......)))))))))...))))). ( -30.80) >DroSec_CAF1 3926 100 + 1 CAUUUUUAUUGGCCAUUUGGCAAAGGCGUCGUGGC-AAACUUUUUCCC-UUAUUCUUGGCGUUGGU---GUGUUUGCCCGCUGCUCUUGGCCAAUACUCAAGUGG (((((.(((((((((...((((...(((....(((-((((.((..(((-........)).)..)).---..))))))))))))))..)))))))))...))))). ( -33.00) >DroEre_CAF1 3919 98 + 1 CAUUUUUAUUGGCCAUUUGGCAAAGGCGUCGUGGC-AAACUUUGUCCC-UCAUUGUUGGCGUUGGU-----UUUUGCCCGCUGCUCUUGGCCAAUAAUCAAGUGG (((((((((((((((...((((...(((....(((-(((..(((.(((-........)).).))).-----.)))))))))))))..))))))))))..))))). ( -31.50) >DroYak_CAF1 4090 98 + 1 CAUUUUUAUUGGCCAUUUGGCAGAGGCGUCGUGGC-AAACUUUGUCCC-UCAUUGUUGGCGUUGGU-----GUUUGUCCGCUGCUCUUGGCCAAUAAUCAAGUGG (((((((((((((((...(((((......((.(((-((((.....(((-........)).).....-----))))))))))))))..))))))))))..))))). ( -33.30) >DroAna_CAF1 3680 97 + 1 CAUUUUUAUUGGCCAUUUGGCAACGACGCUGAGGG-AAACUUUGCGCCAGCAUUGUGGGAGUUGCU-----GCU--GCCACUGGACUUGGCCAAUACUCAAGUGG (((((.(((((((((.(((....)))(((.((((.-...)))))))(((.....((((.(((....-----)))--.)))))))...)))))))))...))))). ( -35.90) >consensus CAUUUUUAUUGGCCAUUUGGCAAAGGCGUCGUGGC_AAACUUUGUCCC_UCAUUGUUGGCGUUGGU_____GUUUGCCCGCUGCUCUUGGCCAAUACUCAAGUGG (((((.(((((((((...(((((((..((....))....)))))))...........((((..(((.........))))))).....)))))))))...))))). (-18.76 = -19.73 + 0.98)


| Location | 7,004,868 – 7,004,966 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.49 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 7004868 98 - 27905053 CCACUUGAGUAUUGGCCAAGAGCAGCGGGCAAAC-----ACCAACGCCAAGAAUAA-GGGACAAAGUUU-GCCACGACGCCUUUGCCAAAUGGCCAAUAAAAAUG .........(((((((((((.((..(((((((((-----.....(.((........-))).....))))-))).))..))))........)))))))))...... ( -27.70) >DroPse_CAF1 4945 100 - 1 CCACUUGAGUAUUGGCCAAGAGCAGUGG-CAGUGGCUGUGGCAGCG---GCAGUGG-AAGAAAAAGUUUCACCAGUGCUUCGUUGCCAAAUGGCCAAUAAAAAUG .........(((((((((...(((((..-.....)))))(((((((---(((.(((-..(((.....))).))).)))..)))))))...)))))))))...... ( -38.30) >DroSec_CAF1 3926 100 - 1 CCACUUGAGUAUUGGCCAAGAGCAGCGGGCAAACAC---ACCAACGCCAAGAAUAA-GGGAAAAAGUUU-GCCACGACGCCUUUGCCAAAUGGCCAAUAAAAAUG .........(((((((((((.((..(((((((((..---.((..............-))......))))-))).))..))))........)))))))))...... ( -26.64) >DroEre_CAF1 3919 98 - 1 CCACUUGAUUAUUGGCCAAGAGCAGCGGGCAAAA-----ACCAACGCCAACAAUGA-GGGACAAAGUUU-GCCACGACGCCUUUGCCAAAUGGCCAAUAAAAAUG ........((((((((((((.((..((((((((.-----.....(.((........-)))......)))-))).))..))))........))))))))))..... ( -26.70) >DroYak_CAF1 4090 98 - 1 CCACUUGAUUAUUGGCCAAGAGCAGCGGACAAAC-----ACCAACGCCAACAAUGA-GGGACAAAGUUU-GCCACGACGCCUCUGCCAAAUGGCCAAUAAAAAUG ........((((((((((...((((.((.(((((-----.....(.((........-))).....))))-)........)).))))....))))))))))..... ( -24.50) >DroAna_CAF1 3680 97 - 1 CCACUUGAGUAUUGGCCAAGUCCAGUGGC--AGC-----AGCAACUCCCACAAUGCUGGCGCAAAGUUU-CCCUCAGCGUCGUUGCCAAAUGGCCAAUAAAAAUG .........(((((((((.......((((--(((-----((((..........))))(((((..((...-..))..))))))))))))..)))))))))...... ( -33.50) >consensus CCACUUGAGUAUUGGCCAAGAGCAGCGGGCAAAC_____ACCAACGCCAACAAUGA_GGGACAAAGUUU_GCCACGACGCCUUUGCCAAAUGGCCAAUAAAAAUG .........(((((((((...((((.((............((................)).....(((.......))).)).))))....)))))))))...... (-16.10 = -15.86 + -0.25)

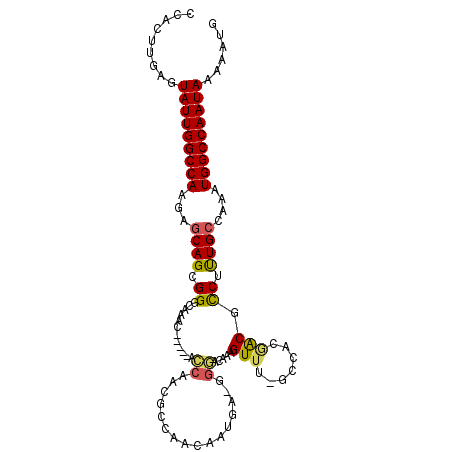

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:01 2006