
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,973,631 – 6,973,750 |
| Length | 119 |
| Max. P | 0.992092 |

| Location | 6,973,631 – 6,973,723 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6973631 92 + 27905053 U--UCCGGACUUUU------------------GAG---ACAGUGCCAAAU-U--UCUUGGCAGUC--GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCGUCAU .--.((((...(((------------------(..---...(((((((.(-(--.((((((((((--(......))))......))))))).)).)))))))....))))))))...... ( -26.80) >DroVir_CAF1 10906 116 + 1 CCAGCCAACCUAUUUGUCUACCCUCUCCCUGGGUGGUUGGUCUGCCAAAAUUUCCUUUGGCAUC----CAUAAACGACAACUGUUGCCAAGGCAAGUGGCAUAAAGCAAAUCGGCAUUGG (((((((......((((((((((.......)))))..(((..(((((((......))))))).)----)).....)))))...((((....)))).)))).....((......))..))) ( -35.40) >DroPse_CAF1 41309 94 + 1 C--UCCAGACUUUUC----------------AGAG---CCUGUGCCAAAU-U--CCUUGGCACUC--GCAUUAACGACAACUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGU .--............----------------.(((---((((((((((.(-(--.((((((((((--(......))).....).))))))).)).)))))))).........))).)).. ( -24.60) >DroEre_CAF1 12970 92 + 1 U--UGCGGAGUUUU------------------GAG---ACAGUGCCAAAU-U--UCUUGGCAGUC--GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCGUCAU .--.((.(((((..------------------..)---)).(((((((.(-(--.((((((((((--(......))))......))))))).)).)))))))........)).))..... ( -28.80) >DroAna_CAF1 10195 91 + 1 U--UUUGCAC---U------------------GAG---GCAGUGCCAAAU-A--UUUUGGCACCCGGGCAUUAAGGACAAAUGUUGCCAAGUAAAUUGGCAUAAAGCAAAUAGGCGUCGU .--..(((.(---(------------------(.(---(...((((((..-.--..)))))))))))))).....(((......((((((.....))))))....((......))))).. ( -27.00) >DroPer_CAF1 42121 94 + 1 C--UCCAGACUUUUC----------------AGAG---CCUGUGCCAAAU-U--CCUUGGCACUC--GCAUUAACGACAACUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGU .--............----------------.(((---((((((((((.(-(--.((((((((((--(......))).....).))))))).)).)))))))).........))).)).. ( -24.60) >consensus C__UCCAGACUUUU__________________GAG___ACAGUGCCAAAU_U__CCUUGGCACUC__GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGU ..........................................((((((........)))))).....((......(((....)))(((((.....))))).............))..... (-14.65 = -14.98 + 0.33)

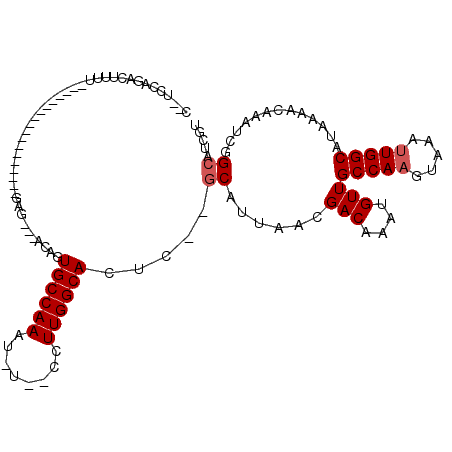
| Location | 6,973,631 – 6,973,723 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6973631 92 - 27905053 AUGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAUUUGUCGUUAAUGC--GACUGCCAAGA--A-AUUUGGCACUGU---CUC------------------AAAAGUCCGGA--A ......(((.((((...((((((((.((.(((((((......(((((....))--)))))))))).--)-).)))))).)).---..)------------------)))....))).--. ( -28.00) >DroVir_CAF1 10906 116 - 1 CCAAUGCCGAUUUGCUUUAUGCCACUUGCCUUGGCAACAGUUGUCGUUUAUG----GAUGCCAAAGGAAAUUUUGGCAGACCAACCACCCAGGGAGAGGGUAGACAAAUAGGUUGGCUGG (((..(((((((((.....(((((.......)))))....(((((.....((----(.((((((((....))))))))..)))...((((.......)))).))))).)))))))))))) ( -39.60) >DroPse_CAF1 41309 94 - 1 ACGAUGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC--GAGUGCCAAGG--A-AUUUGGCACAGG---CUCU----------------GAAAAGUCUGGA--G ...................((((((....(((((((.......((((....))--)).))))))).--.-..))))))((((---((..----------------....))))))..--. ( -28.60) >DroEre_CAF1 12970 92 - 1 AUGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAUUUGUCGUUAAUGC--GACUGCCAAGA--A-AUUUGGCACUGU---CUC------------------AAAACUCCGCA--A .....((.((((((...((((((((.((.(((((((......(((((....))--)))))))))).--)-).)))))).)).---..)------------------)))..)).)).--. ( -28.60) >DroAna_CAF1 10195 91 - 1 ACGACGCCUAUUUGCUUUAUGCCAAUUUACUUGGCAACAUUUGUCCUUAAUGCCCGGGUGCCAAAA--U-AUUUGGCACUGC---CUC------------------A---GUGCAAA--A .....((......))....((((((.....))))))...((((..((....((...((((((((..--.-..))))))))))---...------------------)---)..))))--. ( -23.00) >DroPer_CAF1 42121 94 - 1 ACGAUGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC--GAGUGCCAAGG--A-AUUUGGCACAGG---CUCU----------------GAAAAGUCUGGA--G ...................((((((....(((((((.......((((....))--)).))))))).--.-..))))))((((---((..----------------....))))))..--. ( -28.60) >consensus ACGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC__GAGUGCCAAGA__A_AUUUGGCACAGC___CUC__________________AAAAGUCCGGA__A (((((((......))....((((((.....))))))......)))))...........(((((((......))))))).......................................... (-15.53 = -15.33 + -0.19)


| Location | 6,973,648 – 6,973,750 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6973648 102 + 27905053 AGUGCCAAAU-U--UCUUGGCAGUC--GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCGUCAUCAUCGUUAUUAGC---AUUCCC-CCCUUCCG--------- .(((((((.(-(--.((((((((((--(......))))......))))))).)).)))))))..........((((.......))))......---......-........--------- ( -25.00) >DroVir_CAF1 10946 115 + 1 UCUGCCAAAAUUUCCUUUGGCAUC----CAUAAACGACAACUGUUGCCAAGGCAAGUGGCAUAAAGCAAAUCGGCAUUGGCAUCAUCGUCAGCAUCAGUGUCU-GCCUCAGACCGAGACU ..(((((((......)))))))..----.......(((...((.((((((.((.....((.....))......)).)))))).))..)))((..((.(.((((-(...))))))))..)) ( -30.10) >DroPse_CAF1 41328 88 + 1 UGUGCCAAAU-U--CCUUGGCACUC--GCAUUAACGACAACUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGUCAUCGUUGUUGGC--------------------------- .(((((((..-.--..)))))))..--.......(((((((...((((((.....))))))...........(((...)))...)))))))..--------------------------- ( -26.80) >DroEre_CAF1 12987 103 + 1 AGUGCCAAAU-U--UCUUGGCAGUC--GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCGUCAUCAUCGUUAUUAUC---AUUCCCUCUCUUCCG--------- .(((((((.(-(--.((((((((((--(......))))......))))))).)).)))))))..........((((.......))))......---...............--------- ( -25.00) >DroAna_CAF1 10209 102 + 1 AGUGCCAAAU-A--UUUUGGCACCCGGGCAUUAAGGACAAAUGUUGCCAAGUAAAUUGGCAUAAAGCAAAUAGGCGUCGUCAUUGUUA---UC---GUUCCACCGCUUUUG--------- .(((((((..-.--..))))))).(((........(((((....((((((.....))))))....((......)).......))))).---..---......)))......--------- ( -25.43) >DroPer_CAF1 42140 88 + 1 UGUGCCAAAU-U--CCUUGGCACUC--GCAUUAACGACAACUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGUCAUCGUUGUUGGC--------------------------- .(((((((..-.--..)))))))..--.......(((((((...((((((.....))))))...........(((...)))...)))))))..--------------------------- ( -26.80) >consensus AGUGCCAAAU_U__CCUUGGCACUC__GCAUUAACGACAAAUGUUGCCAAGUAAAUUGGCAUAAAACAAAUCGGCAUCGUCAUCGUUAUUAGC___AUUCCC___CUUC_G_________ ..((((((........)))))).........((((((....(((((((((.....))))))....)))....((.....)).))))))................................ (-14.93 = -15.40 + 0.47)


| Location | 6,973,648 – 6,973,750 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6973648 102 - 27905053 ---------CGGAAGGG-GGGAAU---GCUAAUAACGAUGAUGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAUUUGUCGUUAAUGC--GACUGCCAAGA--A-AUUUGGCACU ---------(....)((-.(..((---.(..........)))..).))...........((((((.((.(((((((......(((((....))--)))))))))).--)-).)))))).. ( -28.10) >DroVir_CAF1 10946 115 - 1 AGUCUCGGUCUGAGGC-AGACACUGAUGCUGACGAUGAUGCCAAUGCCGAUUUGCUUUAUGCCACUUGCCUUGGCAACAGUUGUCGUUUAUG----GAUGCCAAAGGAAAUUUUGGCAGA .(((...(((((...)-))))...)))...((((((..((((((.((......((.....)).....)).))))))...)))))).......----..((((((((....)))))))).. ( -35.00) >DroPse_CAF1 41328 88 - 1 ---------------------------GCCAACAACGAUGACGAUGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC--GAGUGCCAAGG--A-AUUUGGCACA ---------------------------((....(((((..((..((((((.(((........))).....))))))...))..)))))...))--..((((((((.--.-.)))))))). ( -27.90) >DroEre_CAF1 12987 103 - 1 ---------CGGAAGAGAGGGAAU---GAUAAUAACGAUGAUGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAUUUGUCGUUAAUGC--GACUGCCAAGA--A-AUUUGGCACU ---------.((((.(((.((.((---.((.......)).))....))..))).)))).((((((.((.(((((((......(((((....))--)))))))))).--)-).)))))).. ( -27.10) >DroAna_CAF1 10209 102 - 1 ---------CAAAAGCGGUGGAAC---GA---UAACAAUGACGACGCCUAUUUGCUUUAUGCCAAUUUACUUGGCAACAUUUGUCCUUAAUGCCCGGGUGCCAAAA--U-AUUUGGCACU ---------..(((((((.((..(---(.---.........))...))...))))))).((((((.....))))))....................((((((((..--.-..)))))))) ( -24.60) >DroPer_CAF1 42140 88 - 1 ---------------------------GCCAACAACGAUGACGAUGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC--GAGUGCCAAGG--A-AUUUGGCACA ---------------------------((....(((((..((..((((((.(((........))).....))))))...))..)))))...))--..((((((((.--.-.)))))))). ( -27.90) >consensus _________C_GAAG___GGGAAU___GCUAACAACGAUGACGACGCCGAUUUGUUUUAUGCCAAUUUACUUGGCAACAGUUGUCGUUAAUGC__GAGUGCCAAGA__A_AUUUGGCACA ....................................((((((((.((......))....((((((.....))))))....))))))))..........(((((((......))))))).. (-18.83 = -18.38 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:50 2006