
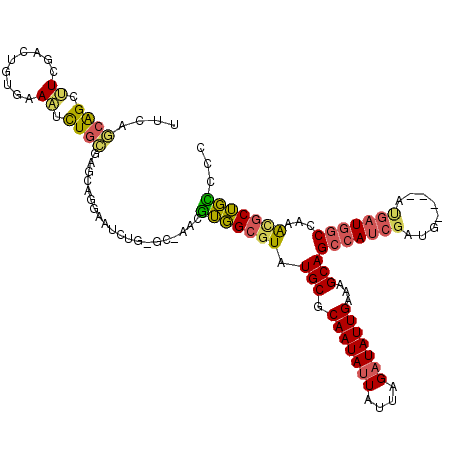
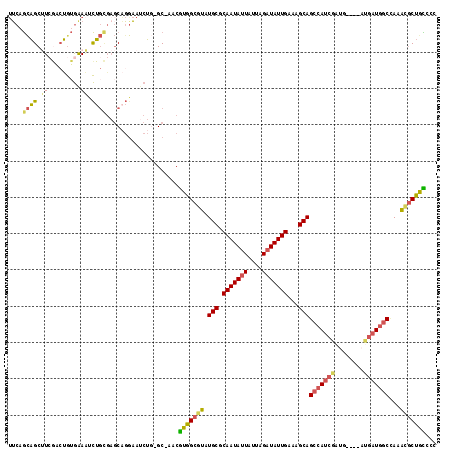
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,969,332 – 6,969,484 |
| Length | 152 |
| Max. P | 0.670212 |

| Location | 6,969,332 – 6,969,444 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.15 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6969332 112 + 27905053 UUCAGCAGCUUCGACUGUGAAAUCUGCGAGCAGGAAUCUG-GC-AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG----AUGAUGGCCAAAUGCUGCCCC ((((.(((......))))))).((((....)))).....(-..-..)(..((((.(((.(((((((....)))))))...)))((((((....----..))))))...))))..)... ( -33.00) >DroSec_CAF1 7099 112 + 1 UUCAGCAGCUUCGACUGUGAAAUCUGCGAGCAGGAAUCUG-GC-AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG----AUGAUGGCCAAACGCUGCCCC ((((.(((......))))))).((((....)))).....(-..-..)(..((((.(((.(((((((....)))))))...)))((((((....----..))))))...))))..)... ( -34.80) >DroSim_CAF1 8949 112 + 1 UUCAGCAGCUUCGACUGUGAAAUCUGCGAGCAGGAAUCUG-GC-AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG----AUGAUGGCCAAACGCUGCCAC ((((.(((......))))))).((((....)))).....(-..-..)(..((((.(((.(((((((....)))))))...)))((((((....----..))))))...))))..)... ( -34.80) >DroEre_CAF1 8901 112 + 1 UUCAGCAGCUUGGACUGUGAAAUCUGCGAGCAGGAAUCUG-GC-AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG----AUGAUGGCUAAGUGCUGCUCC ....((((.((.........)).))))((((((......(-..-..)...((....((.(((((((....)))))))...))(((((((....----..)))))))..)).)))))). ( -33.70) >DroAna_CAF1 7293 103 + 1 UUCUGUAGCAUCAAAUGUGGAUGCU-----CUGAAAUCUC-GC-AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG-------AUGGCCAAA-GCUGUCCC (((.(.((((((.......))))))-----).))).....-..-..((((((...(((.(((((((....)))))))...))))))).))..(-------(((((....-)))))).. ( -28.80) >DroMoj_CAF1 5339 113 + 1 AGCAACGGCAUCAUCCAAAGAGCUUGUGAGCUCGCCGCUCGGCACACAGCGCGUAUGCGCAAUAAUAUUAGAUAUUGAAAGCAGC-AUCGAUGGCUG-CGCUGUCGACGCCCGCU--- (((...(((.((.......(((((....)))))(((....)))..((((((((....)))....................(((((-.......))))-)))))).)).))).)))--- ( -39.10) >consensus UUCAGCAGCUUCGACUGUGAAAUCUGCGAGCAGGAAUCUG_GC_AACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG____AUGAUGGCCAAACGCUGCCCC ....((((.((.........)).))))....................(((((((.(((.(((((((....)))))))...)))(((((((........)))))))...)))))))... (-22.45 = -22.15 + -0.30)

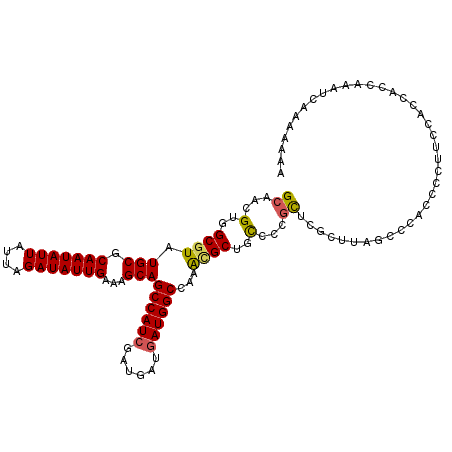
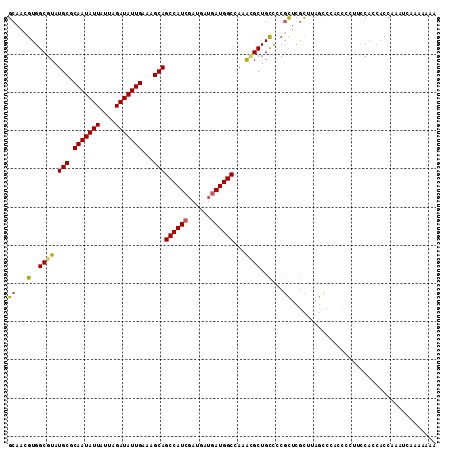
| Location | 6,969,372 – 6,969,484 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.82 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6969372 112 + 27905053 GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAAUGCUGCCCCGCUCGCUUAGCCCAACACUUCCACCACCAAAUCAAAAAAA ((...(..((((.(((.(((((((....)))))))...)))((((((......))))))...))))..)...))...................................... ( -26.40) >DroSec_CAF1 7139 112 + 1 GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAACGCUGCCCCGCUCGCUUAGUCCACCCCUUCCACCACCAAAUCAAAAAAA ((...(..((((.(((.(((((((....)))))))...)))((((((......))))))...))))..)...))...................................... ( -28.20) >DroSim_CAF1 8989 112 + 1 GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAACGCUGCCACGCUCGCUUAGUCCACCCCUUCCACCACCAAAUCAAAAAAA ((..((((((...(((.(((((((....)))))))...)))((((((......))))))........))))))...)).................................. ( -30.10) >DroEre_CAF1 8941 107 + 1 GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCUAAGUGCUGCUCCGCUCGCUCAGCCCACCCUUUCCACAGCCAAA-----AAAA ......((((((((((.(((((((....)))))))...))(((((((......)))))))..))))......(((.....)))..............))))..-----.... ( -27.90) >DroAna_CAF1 7328 95 + 1 GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUG---AUGGCCAAA-GCUGUCCCUUUCCUU----CCUGGCGUUCCCUCACCCAA--------- ((..((((((...(((.(((((((....)))))))...))))))).))..(---(((((....-)))))).........----....))..............--------- ( -22.60) >consensus GCAACGUGGCGUAUGCGCAAUAUUAUUAGAUAUUGAAAGCAGCCAUCGAUGAUGAUGGCCAAACGCUGCCCCGCUCGCUUAGCCCACCCCUUCCACCACCAAAUCAAAAAAA ((...(..((((.(((.(((((((....)))))))...)))((((((......))))))...))))..)...))...................................... (-23.86 = -23.82 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:45 2006