
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,940,091 – 6,940,285 |
| Length | 194 |
| Max. P | 0.929457 |

| Location | 6,940,091 – 6,940,205 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -33.34 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6940091 114 - 27905053 CUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGAGUCCAUGGAAAACUUGGCCAGGCCUCUGAGAAAAUAUGAAUAUGUGCUGAGAUGUAUGUGUA------UGCAUAUGCAUG .((((((.((((((.(((.......))).))))..((((((.((.(((.........))))).))))))..............))))))))(((((((((..------..))))))))). ( -33.00) >DroSim_CAF1 90049 120 - 1 CUUCGGCCACGCCAACUUGAAGUCAAAGAUGGUAAUUAGAGUCCAUGGAAAACUUGGCCAGGCCUCUGAGAAAAUAUGAAUAUGUGCUGAGGUGAAUGUGUACGCAUAUGCAUAUGCAUA (((((((.((((((.(((.......))).))))..((((((.((.(((.........))))).))))))..............)))))))))...(((((((.((....)).))))))). ( -34.10) >DroYak_CAF1 90775 116 - 1 CUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGAGUCCAUGGAAAACUUGGCCAGGCUUCUGAGAAAAUAUGAAUAUGUGCUGAGGUGUAUGUGUA----UAUGCAUAUGCAUA (((((((.((((((.(((.......))).))))..((((((.((.(((.........))))).))))))..............)))))))))(((((((((.----...))))))))).. ( -32.50) >consensus CUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGAGUCCAUGGAAAACUUGGCCAGGCCUCUGAGAAAAUAUGAAUAUGUGCUGAGGUGUAUGUGUA____UAUGCAUAUGCAUA .((((((.((((((.(((.......))).))))..((((((.((.(((.........))))).))))))..............))))))))(((((((((((......))))))))))). (-33.34 = -33.23 + -0.11)

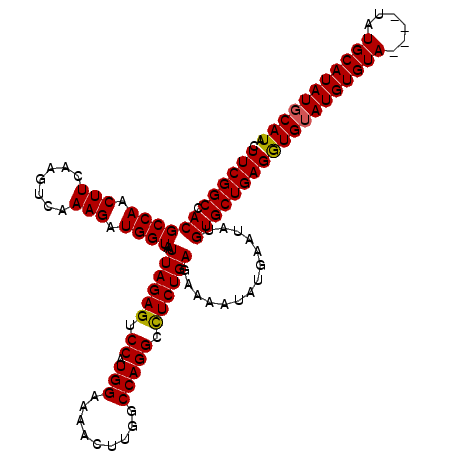
| Location | 6,940,165 – 6,940,285 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -26.95 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6940165 120 - 27905053 AUUCUGCACUCAAAUUGCAUUUUAAAGUAGGCAACAGCAAUCAAGGGAUGUAUUAAGAAAUUAACCCAGGCAGAAAGUUGCUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGA .((((((.(((..((((((((....))).(....).)))))...))).((..((((....))))..)).)))))).((.((....)).))((((.(((.......))).))))....... ( -26.90) >DroSim_CAF1 90129 120 - 1 AUUCUGCACUCAAAUUGCAUUCUAAAGUAGGCAGCAGCAAUCAAGGGCUGUAUUAAGAAAUUAAGCCAGGCAGAAAGUUGCUUCGGCCACGCCAACUUGAAGUCAAAGAUGGUAAUUAGA .((((((........(((........)))(((.(((((........))))).((((....)))))))..)))))).((.((....)).))((((.(((.......))).))))....... ( -33.40) >DroYak_CAF1 90851 120 - 1 AUACUGCACUUAAAUUGCAAUCUUAAGUAGGCAACAGCAAUCGAGGGCUGUAUUAAGAAAUUAACCCAGGCAGAAUGUUGCUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGA ............((((((.(((((((((.(((...((((.((...(.(((..((((....))))..))).).)).))))((....))...))).)))).......))))).))))))... ( -30.51) >consensus AUUCUGCACUCAAAUUGCAUUCUAAAGUAGGCAACAGCAAUCAAGGGCUGUAUUAAGAAAUUAACCCAGGCAGAAAGUUGCUUCGGCCACGCCAACUUCAAGUCAAAGAUGGUAAUUAGA ....((((.......)))).(((((.((.(((...((((((....(.(((..((((....))))..))).).....))))))...)))))((((.(((.......))).))))..))))) (-26.95 = -27.40 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:30 2006