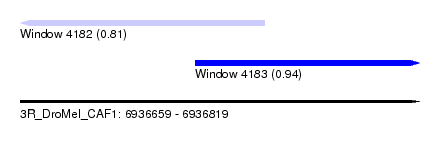
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,936,659 – 6,936,819 |
| Length | 160 |
| Max. P | 0.935706 |

| Location | 6,936,659 – 6,936,757 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6936659 98 - 27905053 AGCUCCU-----------CGGUUCUAUUUGCUGCCCCUGCUGCUUUGGCU---------GCUCCCGGAGUCGCAAGUCAGUCACUUGCAGGAUGCUCUCGUCCUGCAGCUACACACCA .......-----------.(((.......((((.(..(((.(((((((..---------....))))))).))).).))))...((((((((((....))))))))))......))). ( -31.00) >DroPse_CAF1 121823 115 - 1 UGCUUCUACUCCCACCCCUACUCCUAUUU-CUGCUGCUGUUUCUAUGGCUAUUGCUUUUGCUCCUGGAGUCGCA-GUCAGUCACUUGCAGGAUGCUGUCGUCCUGCAGCUACACAC-A .((..........................-(((((((...(((((.((.....((....)).)))))))..)))-).))).....(((((((((....))))))))))).......-. ( -29.00) >DroSec_CAF1 85314 97 - 1 AGCUCCU-----------CGGUUCUAUUUGCUGCCCCUGCUGCUUUGGCU---------GCUCCC-GAGUCGCAAGUCAGUCACUUGCAGGAUGCUCUCGUCCUGCAGCUACACACCA .......-----------.(((.......((.((....)).))..(((((---------((....-.....((((((.....))))))((((((....)))))))))))))...))). ( -29.70) >DroSim_CAF1 86597 97 - 1 AGCUCCU-----------CGGUUCUAUUUGCUGCCCCUGCUGCUUUGGCU---------GCUCCC-GAGUCGCAAGUCAGUCACUUGCAGGAUGCUCUCGUCCUGCAGCUACACACCA .......-----------.(((.......((.((....)).))..(((((---------((....-.....((((((.....))))))((((((....)))))))))))))...))). ( -29.70) >DroYak_CAF1 87302 97 - 1 AGCUCCU-----------CGGUUGUAUUUGCUGCUCCUGCUGCUUUGGCU---------GCUCCC-GAGUCGCAAGUCAGUCACUUGCAGGAUGCUAUCGUCCUGCAGCUACACACCA (((....-----------.(..((.((((((.((((..((.((....)).---------))....-)))).))))))))..)...(((((((((....))))))))))))........ ( -35.50) >consensus AGCUCCU___________CGGUUCUAUUUGCUGCCCCUGCUGCUUUGGCU_________GCUCCC_GAGUCGCAAGUCAGUCACUUGCAGGAUGCUCUCGUCCUGCAGCUACACACCA (((..........................((((....(((.((((.(((..........)))....)))).)))...))))....(((((((((....))))))))))))........ (-22.62 = -23.26 + 0.64)

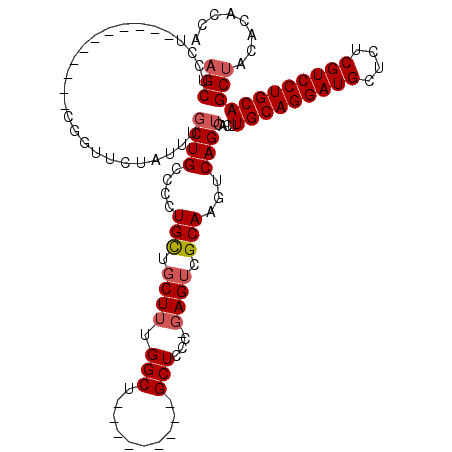
| Location | 6,936,729 – 6,936,819 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -25.11 |
| Energy contribution | -25.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6936729 90 + 27905053 CAGGGGCAGCAAAUAGAACCGAGGAGCUGUAACAACACCUCGCAUCCUUGGCUCUUGCUGUUCGCUGUCAAAAGAGAUUUUUUCAAAUUA (((((((((((...(((.((((((((((((....)))....)).))))))).))))))))))).)))....................... ( -28.30) >DroSec_CAF1 85383 90 + 1 CAGGGGCAGCAAAUAGAACCGAGGAGCUGUAGCAACACCUCGCAUCCUUGGCUCUAGCUGUUCGCUGUCAAAAGAGAUUUUUUCAAAUUA ((((((((((...((((.((((((((((((....)))....)).))))))).))))))))))).)))....................... ( -28.00) >DroSim_CAF1 86666 90 + 1 CAGGGGCAGCAAAUAGAACCGAGGAGCUGUAACAACACCUCGCAUCCUUGGCUGUAGCUGUUCGCUGUCAAAAGAGAUUUUUUCAAAUUA ((((((((((..((((..((((((((((((....)))....)).))))))))))).))))))).)))....................... ( -25.50) >DroYak_CAF1 87371 90 + 1 CAGGAGCAGCAAAUACAACCGAGGAGCUGUAACAACACCUCGCAUCCUUGGCUCUAGCUGUUCGCUGUCAAAAGAGAUUUUUUCAAAUUA ((((((((((........((((((((((((....)))....)).))))))).....))))))).)))....................... ( -23.72) >consensus CAGGGGCAGCAAAUAGAACCGAGGAGCUGUAACAACACCUCGCAUCCUUGGCUCUAGCUGUUCGCUGUCAAAAGAGAUUUUUUCAAAUUA ((((((((((...((((.((((((((((((....)))....)).))))))).))))))))))).)))....................... (-25.11 = -25.68 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:24 2006