
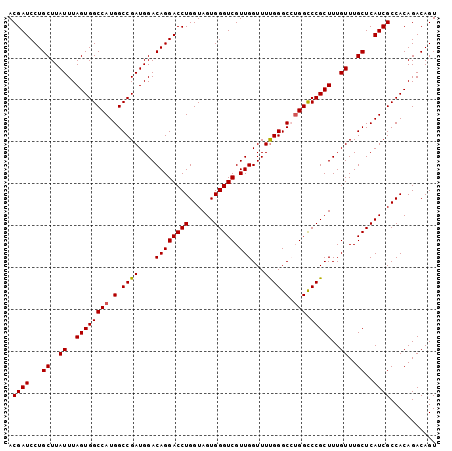
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,932,465 – 6,932,687 |
| Length | 222 |
| Max. P | 0.995013 |

| Location | 6,932,465 – 6,932,585 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -32.52 |
| Energy contribution | -32.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6932465 120 - 27905053 GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUGCCAUGGGCGCAGCUUCAAGGGGGCAGCUAGCUCACUGGCGGGGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ....((((((((.(((((..((((((((.....))))(((((...)))))..))))...(((((.(((((....)))))...)))))))))).)))).))))(((((....))))).... ( -39.20) >DroSec_CAF1 81109 115 - 1 GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUGCCAGGGGCGCAGCUUCUAGGGGGCAG----CUCACUGGCG-GGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ......((.(((((..((((((((..((.(((...((.((((((((((...((((.....)))).)----))).))))))-.)).))).))...))))))))))))).)).......... ( -41.10) >DroSim_CAF1 82400 115 - 1 GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUGCCAGGGGCGCAGCUUGAAGGGGGCAG----CUCACUGGCG-GGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ......((.(((((..((((((((..((.(((...((.((((((((((((..((....))..)).)----))).))))))-.)).))).))...))))))))))))).)).......... ( -39.70) >DroEre_CAF1 83010 115 - 1 GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCCUGCCAGGGGCGCUGCUUCAAGGGGGCAG----CUCUCUGGCA-GGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ......((.(((((..((((((((..((.(((...((((((((((((.(((((((.....))))))----))))))))))-))).))).))...))))))))))))).)).......... ( -52.30) >DroYak_CAF1 83126 115 - 1 GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUGCCAGGGGCGUAGCUUCAAGGGGGCAG----CUCUCUGGCA-GGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ......((.(((((..((((((((..((.(((...((.((((((((((...((((.....)))).)----).))))))))-.)).))).))...))))))))))))).)).......... ( -41.80) >consensus GUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUGCCAGGGGCGCAGCUUCAAGGGGGCAG____CUCACUGGCG_GGCUCUACAUUGAUUUCGCAUUCAUUGGCUGAUGAAGAG ....((((((((.((((((.(((.((((.....)))).(((((((((....((((.....))))......))).))))))...))))))))).)))).))))(((((....))))).... (-32.52 = -32.48 + -0.04)


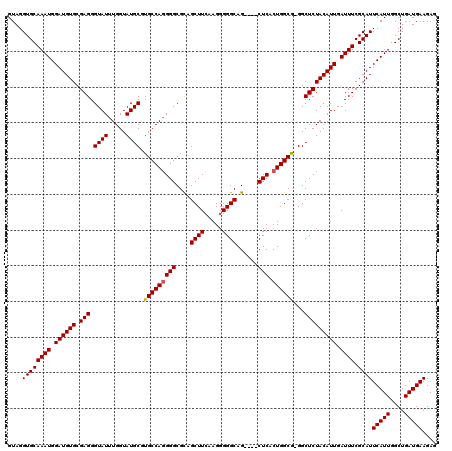
| Location | 6,932,545 – 6,932,661 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 98.10 |
| Mean single sequence MFE | -43.46 |
| Consensus MFE | -42.92 |
| Energy contribution | -43.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6932545 116 - 27905053 GCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUG ((((((...))))...(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))..))... ( -44.64) >DroSec_CAF1 81184 116 - 1 GCGGGCCUGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUG ((((((...))))...(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))..))... ( -44.64) >DroSim_CAF1 82475 116 - 1 GCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUG ((((((...))))...(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))..))... ( -44.64) >DroEre_CAF1 83085 116 - 1 GCGGACCAUGCCCGAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCCUG .(((.......)))..(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))....... ( -38.74) >DroYak_CAF1 83201 116 - 1 GCGGGCCAUGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUG ((((((...))))...(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))..))... ( -44.64) >consensus GCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGUGUAGGUGCAAAUGGAUGUGCGAGGGUAUUUGGUAUGCGUG ((((((...))))...(((((..((((.((((..((((((((.....(((....))).........))))))))..))))(..((......))..)..))))..)))))..))... (-42.92 = -43.12 + 0.20)

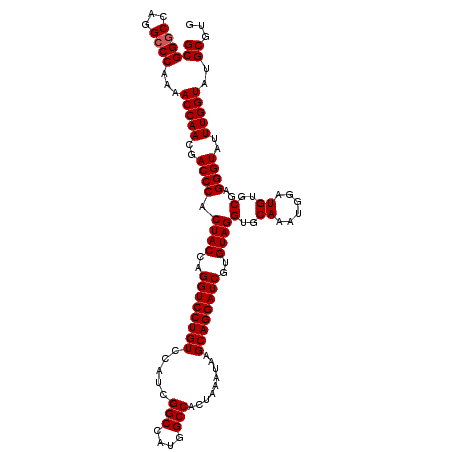
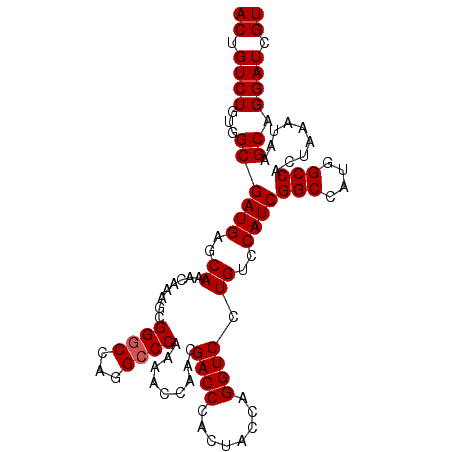
| Location | 6,932,585 – 6,932,687 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 98.24 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6932585 102 + 27905053 ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUUGGGCCUGGCCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..((((((((.((((.(....((((((((......))))).)))....).))))))).)))))..))..))..))))........... ( -36.60) >DroSec_CAF1 81224 102 + 1 ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUUGGGCCAGGCCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..(((((((.(((((.(....((((((((......))))).)))....).))))))).)))))..))..))..))))........... ( -36.60) >DroSim_CAF1 82515 102 + 1 ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUUGGGCCUGGCCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..((((((((.((((.(....((((((((......))))).)))....).))))))).)))))..))..))..))))........... ( -36.60) >DroEre_CAF1 83125 102 + 1 ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUCGGGCAUGGUCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..((((((((((.((((....((((((((......))))).)))...)))).))))).)))))..))..))..))))........... ( -40.30) >DroYak_CAF1 83241 102 + 1 ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUUGGGCAUGGCCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..((((((((((.((((....((((((((......))))).)))...)))).))))).)))))..))..))..))))........... ( -38.20) >consensus ACGAUCCUGCUUAUUUAGUGGCCAUGGCCGAUGGACAGGACCUGGUAGUGGGUCGUUGGUUUUGGGCCUGGCCCGCUUUGUUUGCUCAUCGCCACAGACAGU .((((...((..((..((((((((.(.((((....((((((((......))))).)))...)))).).))).)))))..))..))..))))........... (-33.86 = -33.90 + 0.04)


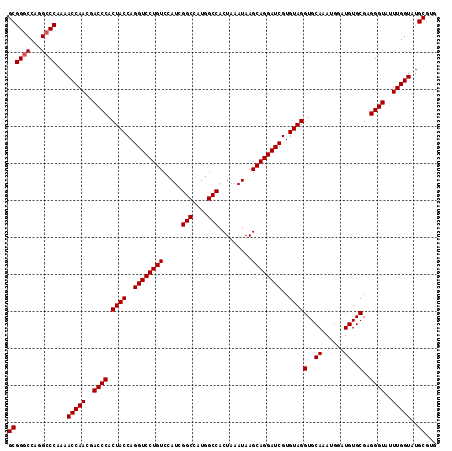
| Location | 6,932,585 – 6,932,687 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 98.24 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6932585 102 - 27905053 ACUGUCUGUGGCGAUGAGCAAACAAAGCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU ((.((((...((((((..((........((((...)))).........((((........)))).))..))))(((....))).........)).)))).)) ( -30.20) >DroSec_CAF1 81224 102 - 1 ACUGUCUGUGGCGAUGAGCAAACAAAGCGGGCCUGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU .((((..(((((.(((.((.......((((((((((.....................))))).)))))......))))).))))).......))))...... ( -32.32) >DroSim_CAF1 82515 102 - 1 ACUGUCUGUGGCGAUGAGCAAACAAAGCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU ((.((((...((((((..((........((((...)))).........((((........)))).))..))))(((....))).........)).)))).)) ( -30.20) >DroEre_CAF1 83125 102 - 1 ACUGUCUGUGGCGAUGAGCAAACAAAGCGGACCAUGCCCGAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU .(((..(((.((.....))......((.((.(((((.((((.......((((........)))).......)))).))))))).))..)))..)))...... ( -29.14) >DroYak_CAF1 83241 102 - 1 ACUGUCUGUGGCGAUGAGCAAACAAAGCGGGCCAUGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU ((.((((...((((((..((........((((...)))).........((((........)))).))..))))(((....))).........)).)))).)) ( -30.20) >consensus ACUGUCUGUGGCGAUGAGCAAACAAAGCGGGCCAGGCCCAAAACCAACGACCCACUACCAGGUCCUGUCCAUCGGCCAUGGCCACUAAAUAAGCAGGAUCGU ((.((((...((((((..((........((((...)))).........((((........)))).))..))))(((....))).........)).)))).)) (-28.66 = -28.86 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:14 2006