
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,921,135 – 6,921,278 |
| Length | 143 |
| Max. P | 0.959534 |

| Location | 6,921,135 – 6,921,252 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6921135 117 + 27905053 UUGCAGUCGUUCGUUUAUGUUUGU-CCUGGUCAGGCCAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG .....((((..(((..((((((((-(((....)))..))))))))...)))...))))......(((((.....)))))...((((((.(((((((((...))))))))))).)))). ( -30.00) >DroSec_CAF1 69776 117 + 1 UUGCAAUCGUUCGUUUAUGUUUGU-CCUGGUCAGGCCAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG ...((((.((......((((((((-(((....)))..)))))))).....)))))).(((....(((((.....)))))......(((.(((((((((...)))))))))))).))). ( -28.70) >DroSim_CAF1 70028 117 + 1 UUGCAAUCGUUCGUUUAUGUUUGU-CCUGGUCAGGCCAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG ...((((.((......((((((((-(((....)))..)))))))).....)))))).(((....(((((.....)))))......(((.(((((((((...)))))))))))).))). ( -28.70) >DroEre_CAF1 71648 117 + 1 UUGCAAUCGUUCGUUUAUGUUUGU-CCUGGUCAGGCCAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG ...((((.((......((((((((-(((....)))..)))))))).....)))))).(((....(((((.....)))))......(((.(((((((((...)))))))))))).))). ( -28.70) >DroYak_CAF1 71635 117 + 1 UUGCAAUCGUUCGUUUAUGUUUGU-CCUGGUCAGGCUAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG ...((((.((......((((((((-(((....)))..)))))))).....)))))).(((....(((((.....)))))......(((.(((((((((...)))))))))))).))). ( -28.70) >DroAna_CAF1 67712 118 + 1 UUACAAUGGUUCGUUUAUGUUUGUGCCUGGUCAGGCAAUAAAUAUUUUAUGCAUUGACACUUUUUCGGCACUAAGCCAAUAAUACGCACACAACAGUGACUCAUUGUUGUUGCCGUGG ...(((((...(((..((((((((((((....)))).))))))))...)))))))).(((......(((.....)))........(((.(((((((((...)))))))))))).))). ( -34.70) >consensus UUGCAAUCGUUCGUUUAUGUUUGU_CCUGGUCAGGCCAUAAAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGG .((((.......(((((((((((........))))).))))))......))))....(((....(((((.....)))))......(((.(((((((((...)))))))))))).))). (-27.68 = -27.74 + 0.06)

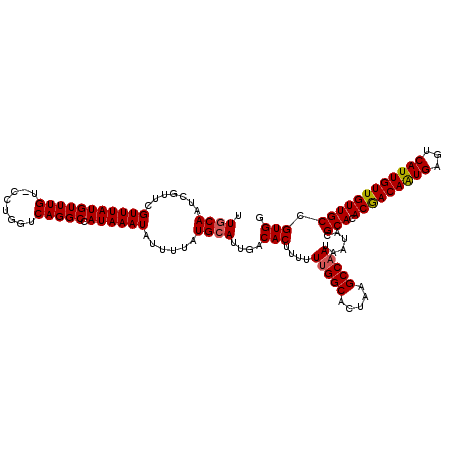
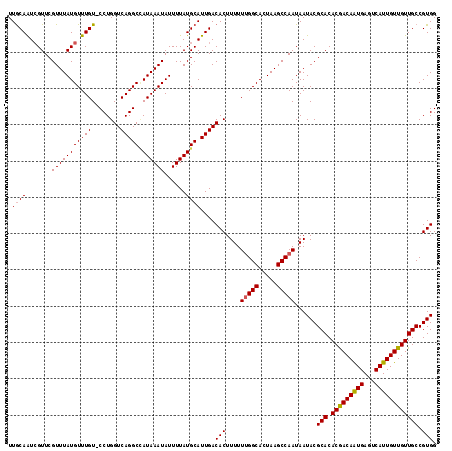
| Location | 6,921,135 – 6,921,252 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6921135 117 - 27905053 CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUGGCCUGACCAGG-ACAAACAUAAACGAACGACUGCAA .(((.(((((.((((((...)))))).)).)))......(((((.....)))))....)))....((((.......((((((.(((....)))-.....))))))........)))). ( -26.16) >DroSec_CAF1 69776 117 - 1 CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUGGCCUGACCAGG-ACAAACAUAAACGAACGAUUGCAA .(((.(((((.((((((...)))))).)).)))......(((((.....)))))....)))....((((.......((((((.(((....)))-.....))))))........)))). ( -26.86) >DroSim_CAF1 70028 117 - 1 CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUGGCCUGACCAGG-ACAAACAUAAACGAACGAUUGCAA .(((.(((((.((((((...)))))).)).)))......(((((.....)))))....)))....((((.......((((((.(((....)))-.....))))))........)))). ( -26.86) >DroEre_CAF1 71648 117 - 1 CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUGGCCUGACCAGG-ACAAACAUAAACGAACGAUUGCAA .(((.(((((.((((((...)))))).)).)))......(((((.....)))))....)))....((((.......((((((.(((....)))-.....))))))........)))). ( -26.86) >DroYak_CAF1 71635 117 - 1 CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUAGCCUGACCAGG-ACAAACAUAAACGAACGAUUGCAA .....((((...(((((...)))))((((...(((....(((((.....)))))....(.((((..((((((.....)))).)).)))))...-..........))).)))))))).. ( -23.50) >DroAna_CAF1 67712 118 - 1 CCACGGCAACAACAAUGAGUCACUGUUGUGUGCGUAUUAUUGGCUUAGUGCCGAAAAAGUGUCAAUGCAUAAAAUAUUUAUUGCCUGACCAGGCACAAACAUAAACGAACCAUUGUAA ....(....).((((((..((..((((.(((((......(((((.....)))))....(.((((..(((............))).)))))..))))))))).....))..)))))).. ( -28.20) >consensus CCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUUUAUGGCCUGACCAGG_ACAAACAUAAACGAACGAUUGCAA .(((.(((((.((((((...)))))).)).)))......(((((.....)))))....)))....((((.......((((((.(((....)))......))))))........)))). (-23.00 = -23.23 + 0.22)

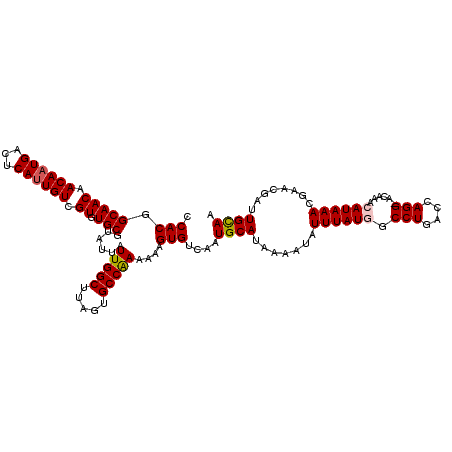
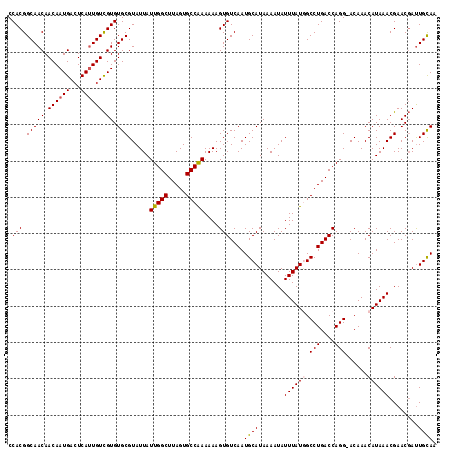
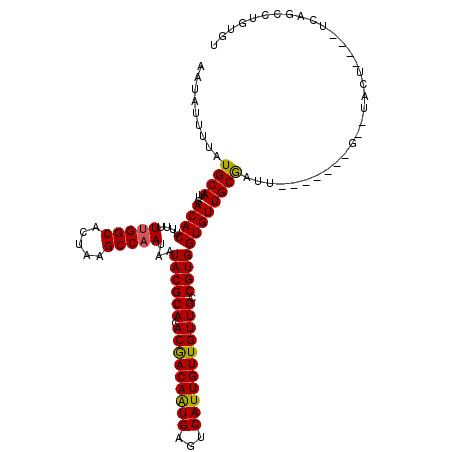
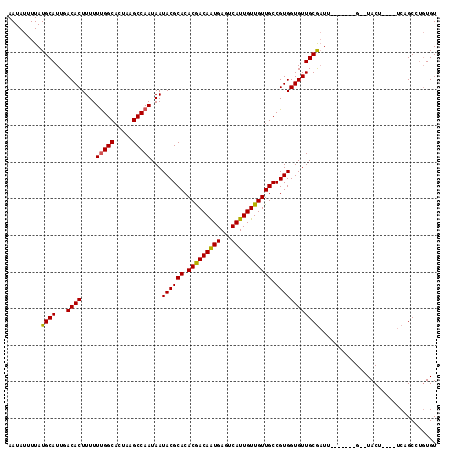
| Location | 6,921,174 – 6,921,278 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6921174 104 + 27905053 AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU-------G--UACU----UCUGCCUGUGU .........((((...((((....(((((.....)))))...((((((.(((((((((...))))))))))).))))))))))))...-------.--....----........... ( -24.90) >DroSec_CAF1 69815 106 + 1 AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU-------GUGUACU----UCUGCCUGUGU ..........(((...((((....(((((.....)))))((((((.(((.((((((..(....)..))))))..))))))))).....-------))))...----..)))...... ( -26.50) >DroSim_CAF1 70067 106 + 1 AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU-------GUGUACU----UCUGCCUGUGU ..........(((...((((....(((((.....)))))((((((.(((.((((((..(....)..))))))..))))))))).....-------))))...----..)))...... ( -26.50) >DroEre_CAF1 71687 104 + 1 AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU-------G--UACU----UCAGCCUGUGU .........((((...((((....(((((.....)))))...((((((.(((((((((...))))))))))).))))))))))))...-------.--....----........... ( -24.90) >DroYak_CAF1 71674 104 + 1 AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU-------G--UACU----UCAGCCUGUGU .........((((...((((....(((((.....)))))...((((((.(((((((((...))))))))))).))))))))))))...-------.--....----........... ( -24.90) >DroAna_CAF1 67752 117 + 1 AAUAUUUUAUGCAUUGACACUUUUUCGGCACUAAGCCAAUAAUACGCACACAACAGUGACUCAUUGUUGUUGCCGUGGUGUUGCACUUGCUGUUUGUGUACUUGUUUCAGCCGCUGU ..........(((...((((.....(((((....((.((((.((((((.(((((((((...))))))))))).)))).))))))...)))))...))))...)))............ ( -28.60) >consensus AAUAUUUUAUGCAUUGACACUUUUUUGGCACUAAGCCAAUAAUACGCACACGACAAUGAGUCAUUGUUGUUGCCGUGGUGUUGCGAUU_______G__UACU____UCAGCCUGUGU .........((((...((((....(((((.....)))))...((((((.(((((((((...))))))))))).))))))))))))................................ (-24.76 = -24.52 + -0.25)

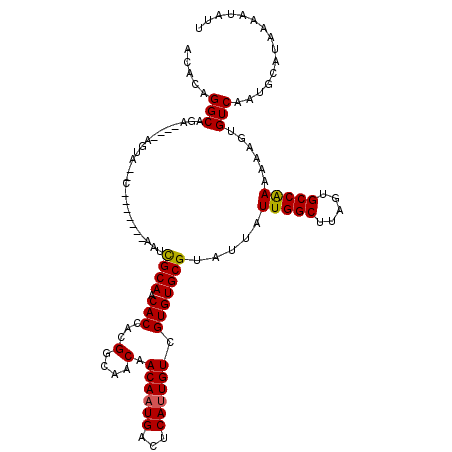
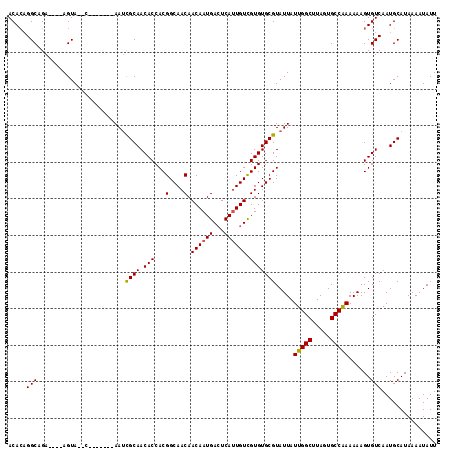
| Location | 6,921,174 – 6,921,278 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6921174 104 - 27905053 ACACAGGCAGA----AGUA--C-------AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU ......(((..----..((--(-------(((((((.(((...(....)......(((.....)))))))))).))).(((((.....)))))....)))....))).......... ( -24.00) >DroSec_CAF1 69815 106 - 1 ACACAGGCAGA----AGUACAC-------AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU ......(((..----.(.((((-------(((((((.(((...(....)......(((.....)))))))))).))).(((((.....)))))....)))))..))).......... ( -26.60) >DroSim_CAF1 70067 106 - 1 ACACAGGCAGA----AGUACAC-------AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU ......(((..----.(.((((-------(((((((.(((...(....)......(((.....)))))))))).))).(((((.....)))))....)))))..))).......... ( -26.60) >DroEre_CAF1 71687 104 - 1 ACACAGGCUGA----AGUA--C-------AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU .....(((..(----(((.--(-------(((((((.(((...(....)......(((.....))))))))))....))))))))...)))......((((....))))........ ( -22.60) >DroYak_CAF1 71674 104 - 1 ACACAGGCUGA----AGUA--C-------AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU .....(((..(----(((.--(-------(((((((.(((...(....)......(((.....))))))))))....))))))))...)))......((((....))))........ ( -22.60) >DroAna_CAF1 67752 117 - 1 ACAGCGGCUGAAACAAGUACACAAACAGCAAGUGCAACACCACGGCAACAACAAUGAGUCACUGUUGUGUGCGUAUUAUUGGCUUAGUGCCGAAAAAGUGUCAAUGCAUAAAAUAUU ...(((..(((.((..((((((((.(((...(((......)))(....)............)))))))))))......(((((.....)))))....)).))).))).......... ( -29.90) >consensus ACACAGGCAGA____AGUA__C_______AAUCGCAACACCACGGCAACAACAAUGACUCAUUGUCGUGUGCGUAUUAUUGGCUUAGUGCCAAAAAAGUGUCAAUGCAUAAAAUAUU .....(((........................((((.(((...(....).((((((...)))))).))))))).....(((((.....)))))......)))............... (-20.61 = -20.50 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:39:03 2006