
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,906,576 – 6,906,684 |
| Length | 108 |
| Max. P | 0.733675 |

| Location | 6,906,576 – 6,906,684 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -16.93 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6906576 108 + 27905053 G--CCAAACCG-----AAC-----CGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUAACAUGAAAUGUGCAUCAUC .--........-----...-----.(((.((((((.....((((............))))(((((((........))..((((((.....))))))..)))))....)))))).)))... ( -15.90) >DroPse_CAF1 76355 113 + 1 A--CCAAACCGGGUAAAAC-----CGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAAUAACAUGAAAUGUGCAUCAUC .--......(((......)-----))..........((((((((...((((....)))).((((.((........))..((((((.....))))))...)))).....)))))))).... ( -18.70) >DroWil_CAF1 65387 109 + 1 GAGAC-AGUCC-----AAC-----CGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAAAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUACCAUGAAAUGUGCAGCAUC .....-.....-----...-----.............(((((((...((((.....(((((((......)))........(((((.....)))))))))...))))..)))))))..... ( -12.80) >DroYak_CAF1 56616 113 + 1 G--CCAAACCG-----AACCGUACCGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUAACAUGAAAUGUGCAUCAUC .--........-----....((((...((((..(((((((((((............))))((((((......................)))))).)))))))..))))..))))...... ( -17.05) >DroMoj_CAF1 59416 102 + 1 G--GCA-----------AG-----CGAUUCGCAGUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUAACAUGAAAUGUGCAUCAUC .--...-----------.(-----((...)))....((((((((...((((....)))).(((((((........))..((((((.....))))))..))))).....)))))))).... ( -17.50) >DroPer_CAF1 77222 113 + 1 A--CCAAACCGGGUAAAAC-----CGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUAACAUGAAAUGUGCAUCAUC .--......(((......)-----))..........((((((((...((((....)))).(((((((........))..((((((.....))))))..))))).....)))))))).... ( -19.60) >consensus G__CCAAACCG_____AAC_____CGAUUCACAUUAAUGCGCAUAAAUAUGAAAUCAUGCGUUAAGAUCAACAAUUCAACAAUUAAAACUUAAUUGCAUUAACAUGAAAUGUGCAUCAUC .........................(((.((((((...((((((............)))))).................((((((.....))))))...........)))))).)))... (-14.32 = -14.52 + 0.19)
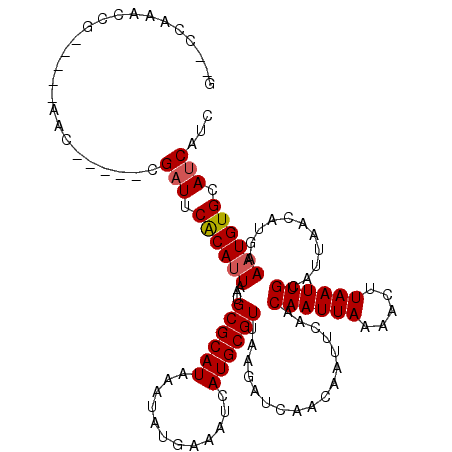
| Location | 6,906,576 – 6,906,684 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6906576 108 - 27905053 GAUGAUGCACAUUUCAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG-----GUU-----CGGUUUGG--C ..(((.((....(((((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))))...-----)))-----))......--. ( -26.00) >DroPse_CAF1 76355 113 - 1 GAUGAUGCACAUUUCAUGUUAUUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG-----GUUUUACCCGGUUUGG--U ..(((((((((.((((.((((..((......))..))))...)))).)))..........((((((........))))))))))))....((((((-----(......)))))))..--. ( -27.20) >DroWil_CAF1 65387 109 - 1 GAUGCUGCACAUUUCAUGGUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUUUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG-----GUU-----GGACU-GUCUC (((..(.(((..(((((..((((((...............((((((........))))))((((((........))))))))))))..)))))...-----).)-----).)..-))).. ( -24.50) >DroYak_CAF1 56616 113 - 1 GAUGAUGCACAUUUCAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCGGUACGGUU-----CGGUUUGG--C ((...((.((..(((((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))))...)).))..)-----).......--. ( -27.40) >DroMoj_CAF1 59416 102 - 1 GAUGAUGCACAUUUCAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUACUGCGAAUCG-----CU-----------UGC--C ..(((((((((.((((.(((((.((......)).)))))...)))).)))..........((((((........))))))))))))..((((....-----.)-----------)))--. ( -21.90) >DroPer_CAF1 77222 113 - 1 GAUGAUGCACAUUUCAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG-----GUUUUACCCGGUUUGG--U .............((((((((((((..(((((..(((((.........))))).))))).((((((........))))))))))))))))))((((-----(......)))))....--. ( -27.60) >consensus GAUGAUGCACAUUUCAUGUUAAUGCAAUUAAGUUUUAAUUGUUGAAUUGUUGAUCUUAACGCAUGAUUUCAUAUUUAUGCGCAUUAAUGUGAAUCG_____GUU_____CGGUUUGG__C ............(((((((((((((..(((((..(((((.........))))).))))).((((((........)))))))))))))))))))........................... (-21.62 = -22.15 + 0.53)

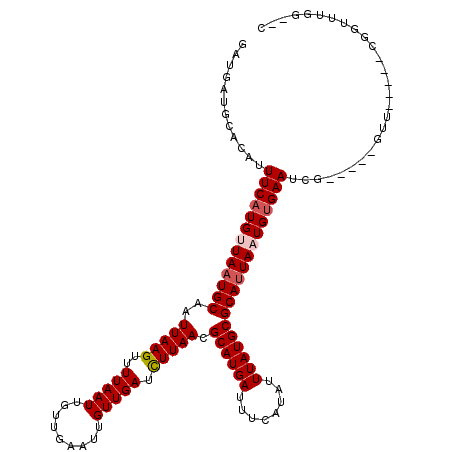
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:52 2006