| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,900,387 – 6,900,483 |
| Length | 96 |
| Max. P | 0.550270 |

| Location | 6,900,387 – 6,900,483 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.79 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
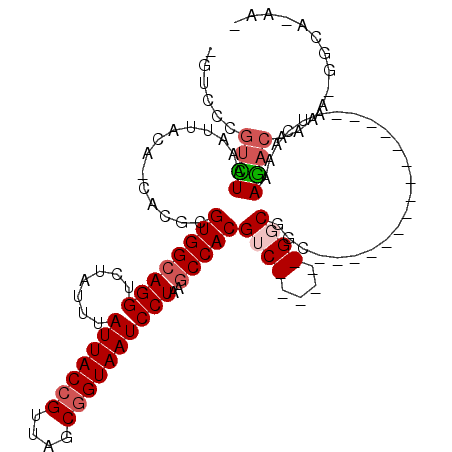
>3R_DroMel_CAF1 6900387 96 + 27905053 -GGCCCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUC------GGCGGC---------------AUUGGAAGACAAAA-GGCAAAAG -.(((.((((.(((..(.-(.(((((((((((.......(((((((....))))))))))..)))))).)------)).)..---------------)))...))))....-)))..... ( -29.81) >DroPse_CAF1 68951 100 + 1 -GUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCGGCGGC---------------AUCAAAAGACAAAGAGGUA---- -.((((((((........((.(((((((((((.......(((((((....))))))))))..)))))).))))((.....))---------------......))))...))))..---- ( -31.31) >DroWil_CAF1 58423 111 + 1 CGCCUCGUCUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACAGUUAACGGUAAUCCUAAGCCACGAC------GGCGGCGGUGCCAACGUCAGCAUCA-AAGACAAAA-GGCAAAAA .((((.((((........-..(((((((((((.......(((((.(....).))))))))..)))))).)------)((((((.......)))).))....-.))))...)-)))..... ( -32.21) >DroMoj_CAF1 54392 96 + 1 -GUCCCCUUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGAC------GGCGCC---------------GAGCUGAAACAAAA-GCAAUAAG -.................-...((((((((((.......(((((((....))))))))))..)))))..(------(((...---------------..))))........-))...... ( -25.61) >DroAna_CAF1 47024 96 + 1 -GGCCCGGUUAAAUUACA-CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAUCCACGUC------GGCGGC---------------AUCAAAAGACAAAA-GGCACAAA -.(((..(((........-(.(((((((.(((.......(((((((....))))))))))...))))).)------)..)..---------------.......)))....-)))..... ( -22.94) >DroPer_CAF1 69735 100 + 1 -GUCUCGUCUAAAUUACAGCACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUCGGCGUCGGCGGC---------------AUCAAAAGACAAAGAGGUA---- -.((((((((........((.(((((((((((.......(((((((....))))))))))..)))))).))))((.....))---------------......))))...))))..---- ( -31.31) >consensus _GUCCCGUCUAAAUUACA_CACGCGUGGCAGGUCUAUUUAUUACCGUUAGCGGUAAUCCUAAGCCACGUC______GGCGGC_______________AUCAAAAGACAAAA_GGCA_AA_ ......((((..............((((((((.......(((((((....))))))))))..)))))((((....))))........................))))............. (-17.99 = -18.79 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:47 2006