
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,872,479 – 6,872,583 |
| Length | 104 |
| Max. P | 0.650290 |

| Location | 6,872,479 – 6,872,583 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -3.78 |
| Energy contribution | -4.62 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6872479 104 + 27905053 GUU-------GGCAAUGAGGCUUUUUAGCUUACUUCAAGGUGAAGGUUGCUUACUUAAGGAGUUAUCCUA--CUUGUAUUCCUGUAUUUCU-UGGAAUGCCAAAUUGUUUUCUU ((.-------((((((.(((((....))))).((((.....)))))))))).))...((((....)))).--...(((((((.........-.))))))).............. ( -26.30) >DroSec_CAF1 26734 104 + 1 GUU-------GGCAAUAAGGCUUUUUGGUU-UAUUC-UGGUGAAGGUUGCUCACUUAAGGAGUUAUCUUAUGCUUGUAUUCCUGUAUUUCU-UGGAAAGCCAAAUUGUUUUCUU (..-------((((((..((((((((((..-(((..-.(((((((((.((((.(....))))).))))).)))).)))..)).........-.))))))))..))))))..).. ( -24.90) >DroSim_CAF1 27034 89 + 1 GUU-------GGC---------------UU-ACUUC-UGGUGAAGGUUGCUCACUUAAGGAGUUAUCUUAUGCUUGUAUUCCUGUAUUUCU-UGGAAGUCCAAAUUGUUUUCUU .((-------((.---------------..-(((((-..(..(((((.((((.(....))))).)))))((((..........))))...)-..)))))))))........... ( -17.10) >DroEre_CAF1 27685 103 + 1 GUU-------GGCAAUGAGGCUUUUGGCCUUAACUC-UGUCGCUAUUUUCUCACUUAAGGAGUAAUCUU--GUUUGUACUUCUGUAUUUCU-UGAAAGCCCAAACUGUUUUUUU (((-------(((..((((((.....))))))....-.................((((((((((.....--((....)).....)))))))-)))..)))..)))......... ( -20.70) >DroYak_CAF1 27399 111 + 1 GUUGGCAAUUGGCAAUGAGGCUUUUUGCCUUACCUC-UGGCGCAGGUUGCUCACUUAAGGCGUUAUCUU--GCUGGUAUUCCUGUAUUUCCAAGAAAGCCCAAAUUGUUUUUUU ((.((((((..((..((((((.....))))))((..-.)).))..)))))).))....((.(((.((((--(..(((((....)))))..))))).)))))............. ( -31.10) >consensus GUU_______GGCAAUGAGGCUUUUUGCCUUACUUC_UGGUGAAGGUUGCUCACUUAAGGAGUUAUCUUA_GCUUGUAUUCCUGUAUUUCU_UGGAAGCCCAAAUUGUUUUCUU ..........((((((..((.....................................((((((...(........).))))))...((((...))))..))..))))))..... ( -3.78 = -4.62 + 0.84)



| Location | 6,872,479 – 6,872,583 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.95 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -2.42 |
| Energy contribution | -1.90 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6872479 104 - 27905053 AAGAAAACAAUUUGGCAUUCCA-AGAAAUACAGGAAUACAAG--UAGGAUAACUCCUUAAGUAAGCAACCUUCACCUUGAAGUAAGCUAAAAAGCCUCAUUGCC-------AAC .......((((..(((.((((.-.........))))(((...--.((((....))))...)))(((...((((.....))))...))).....)))..))))..-------... ( -18.90) >DroSec_CAF1 26734 104 - 1 AAGAAAACAAUUUGGCUUUCCA-AGAAAUACAGGAAUACAAGCAUAAGAUAACUCCUUAAGUGAGCAACCUUCACCA-GAAUA-AACCAAAAAGCCUUAUUGCC-------AAC .......((((..((((((...-........((((..................))))...(((((.....)))))..-.....-......))))))..))))..-------... ( -14.57) >DroSim_CAF1 27034 89 - 1 AAGAAAACAAUUUGGACUUCCA-AGAAAUACAGGAAUACAAGCAUAAGAUAACUCCUUAAGUGAGCAACCUUCACCA-GAAGU-AA---------------GCC-------AAC ..........(((((....)))-)).......((..(((.....((((.......)))).(((((.....)))))..-...))-).---------------.))-------... ( -12.70) >DroEre_CAF1 27685 103 - 1 AAAAAAACAGUUUGGGCUUUCA-AGAAAUACAGAAGUACAAAC--AAGAUUACUCCUUAAGUGAGAAAAUAGCGACA-GAGUUAAGGCCAAAAGCCUCAUUGCC-------AAC .........(((((..((((..-(....)...))))..)))))--..............(((((.....((((....-..)))).(((.....))))))))...-------... ( -16.10) >DroYak_CAF1 27399 111 - 1 AAAAAAACAAUUUGGGCUUUCUUGGAAAUACAGGAAUACCAGC--AAGAUAACGCCUUAAGUGAGCAACCUGCGCCA-GAGGUAAGGCAAAAAGCCUCAUUGCCAAUUGCCAAC .......((((((((...((((((......))))))..)))((--((......(((((..(((((....)).)))..-))))).((((.....))))..)))).)))))..... ( -26.50) >consensus AAGAAAACAAUUUGGCCUUCCA_AGAAAUACAGGAAUACAAGC_UAAGAUAACUCCUUAAGUGAGCAACCUUCACCA_GAAGUAAGCCAAAAAGCCUCAUUGCC_______AAC ..........((((...((((...........))))..))))........................................................................ ( -2.42 = -1.90 + -0.52)

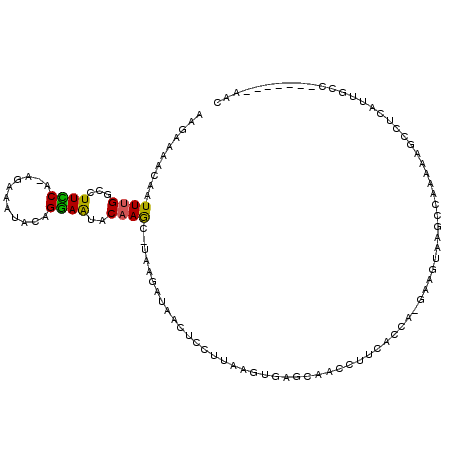
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:31 2006