
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,871,652 – 6,871,856 |
| Length | 204 |
| Max. P | 0.872140 |

| Location | 6,871,652 – 6,871,753 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6871652 101 - 27905053 UUGAUUAAAUUGUUGUGCUG----UGCUGGUGGAGCUCCUUCUGCU-UUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA .........(..(((..(.(----(((((.(..(((.......)))-..).)...(((((((((...((.....))..)))))))))))))))..)))..)..... ( -32.90) >DroSec_CAF1 25906 101 - 1 UUGAUUAAAUUGUUGUGCUG----UGCUGGUGGAGUUCUGGCUGCU-UUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA .........(..(((..(.(----((((...........(((.((.-...)).)))((((((((...((.....))..)))))))).))))))..)))..)..... ( -33.30) >DroSim_CAF1 26204 101 - 1 UUGAUUAAAUUGUUGUGCUG----UGCUGGUGGAGCUCUGGCUGCU-UUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA ....(((.(((((((((((.----.((..(.(.(((....))).).-)..))...(((((((((...((.....))..))))))))))))))).)))))...))). ( -33.60) >DroEre_CAF1 26875 101 - 1 UUGAUUAAAUUGUUGUGCUG----UGCUGGUGGAGCUGCUUCUGCU-UUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA .........(..(((..(.(----((((((..(....((....)).-....)..))((((((((...((.....))..)))))))).))))))..)))..)..... ( -33.80) >DroYak_CAF1 26549 101 - 1 UUGAUUAAAUUGUUGUGCUG----UGCUGGUGGAGCUGCUUCUGCU-UUAGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA .........(..(((..(.(----(((((.((((((.......)))-))).)...(((((((((...((.....))..)))))))))))))))..)))..)..... ( -34.10) >DroAna_CAF1 25153 106 - 1 UUGAUUAAAUUGUUGUGCUGCUACUGAUGCUUCGGCUCCUUCUGCUUUUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAACAAAGCACGUGCAGUGAGUAAA .........(..(((..(((((...((..(..((((.((..........))..))))...)...))(((((((....)))))))...)))).)..)))..)..... ( -22.80) >consensus UUGAUUAAAUUGUUGUGCUG____UGCUGGUGGAGCUCCUUCUGCU_UUGGCUGCCGCUUGAUUUCUUGAUUGCCAUUAAUCAAGCGAGCACGUGCAGUGAGUGAA ....(((.(((((((((((......(((((...(((.......))).)))))...(((((((((...((.....))..))))))))))))))).)))))...))). (-26.47 = -26.75 + 0.28)


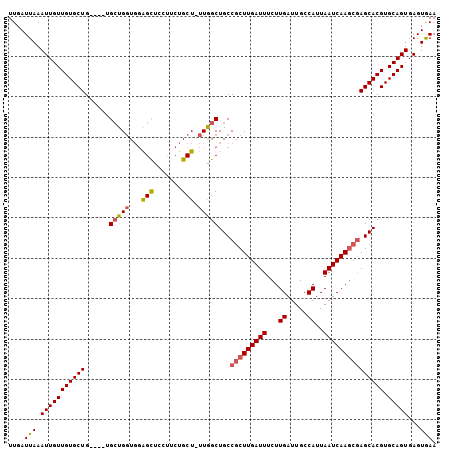
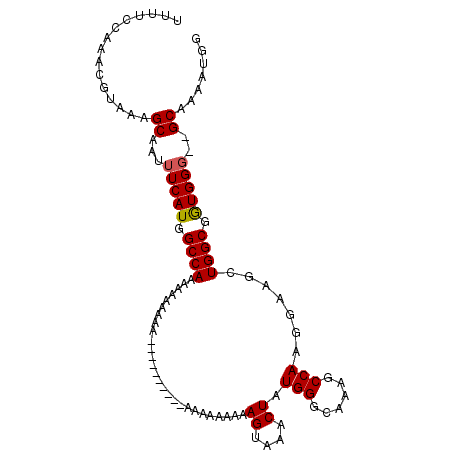
| Location | 6,871,753 – 6,871,856 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -15.27 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6871753 103 + 27905053 UUUUCCAAACGUAAAGCAAUUUCAUGGCCAAAAUAAAAAUAAGAAAAGAAAAAAAAAAGUAAACUAUGGGCAAAGCCAAGGAAGCUGGCGGUGG---GCAAAAUGG ....(((..((((((....))).)))(((............................((....)).....((..((((.(....)))))..)))---))....))) ( -15.00) >DroEre_CAF1 26976 96 + 1 UUUUCGAAACGUAAAGCAAUUUCAUGGCCAAAAAAAAAA----------CCAAAAAAAGUAAACUAUGGGUAAAGCCAAGGAAGCUGGCGGUGGGUAGCAAAAUGG ...............((...(((((.((((........(----------(........))...((.(((......))).))....)))).)))))..))....... ( -15.60) >DroYak_CAF1 26650 95 + 1 UUUUCCAAACGUAAAGCAAUUUCAUGGCCAAAAAAAAGA----------GAGAGAAAAGUAAACUAUGGGGAAAGCCAAGAAAGCUGGCGAUGGG-GGCAAAAUGG ...............((..(..(((.((((.........----------........((....)).(((......))).......)))).)))..-)))....... ( -15.20) >consensus UUUUCCAAACGUAAAGCAAUUUCAUGGCCAAAAAAAAAA__________AAAAAAAAAGUAAACUAUGGGCAAAGCCAAGGAAGCUGGCGGUGGG__GCAAAAUGG ...............((...(((((.((((...........................((....)).(((......))).......)))).)))))..))....... (-14.69 = -14.80 + 0.11)
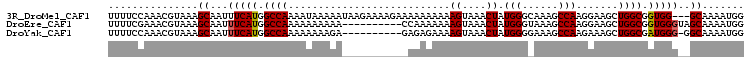

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:30 2006