
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,833,157 – 6,833,340 |
| Length | 183 |
| Max. P | 0.911609 |

| Location | 6,833,157 – 6,833,271 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.71 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6833157 114 - 27905053 CAAUGAUAAUGUCAUACCGAUUUCACUACAUAUCCAACCC-----AGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAAGCCAAAGGGUUGCUUCCAAAUCAGAUGGAAUGGGUAAAU ...((((...))))((((.((((((((.......((((((-----......((((((...))))))..(((((.....)))))...))))))..........)).)))))).))))... ( -29.43) >DroSec_CAF1 92190 114 - 1 CAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC-----AGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAAGCCAAAGGGUUGCUUCCAAAUCAGAUGGAAUGGGUAAAU ...((((...))))((((.((((((((...(((.......-----))).(((..(((((((....)))(((((.....)))))...))))..))).......)).)))))).))))... ( -29.70) >DroSim_CAF1 98504 114 - 1 CAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC-----AGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAAGCCAAAGGGUUGCUUCCAAAUCAGAUGGAAUGGGUAAAU ...((((...))))((((.((((((((...(((.......-----))).(((..(((((((....)))(((((.....)))))...))))..))).......)).)))))).))))... ( -29.70) >DroEre_CAF1 77276 114 - 1 CAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC-----AGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAACCCAAAGGGUUGCCUUCAAAUCAGAUGGAACGGGUAAAU ..(((((...))))).(((...............((((((-----......((((((...))))))(.((((......)))))...))))))((.((......)).))..)))...... ( -25.90) >DroYak_CAF1 90838 119 - 1 CAAUGAUAAUGUCAUACCAAUUUCACUACAUCUACGACCCAGACCAGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAACCCAAAGGGUUGCUUCCAAAUCAGAUGGAAUGGGUAAAU ..(((((...))))).....................(((((..(((......(((.(((((....))))).......((((((...))))))...))).......)))..))))).... ( -30.12) >consensus CAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC_____AGACAAGUGGCCUGCGGUUACGCGGUUUAAUACAAGCCAAAGGGUUGCUUCCAAAUCAGAUGGAAUGGGUAAAU ...((((...))))((((.((((((((.......((((((...........((((((...))))))..(((((.....)))))...))))))..........)).)))))).))))... (-25.27 = -25.71 + 0.44)


| Location | 6,833,231 – 6,833,340 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6833231 109 - 27905053 AGAAGUCGCUCACAGGAGCAUAUAG------CACACAUACA-----UCAUGCAUUGCUGGAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCGAUUUCACUACAUAUCCAACCC .((((((((((....))))......------...((((..(-----(((((((((((((((....)))))).....))))).))))).))))......))))))................ ( -30.50) >DroSec_CAF1 92264 110 - 1 AGAAGUCGUUCACAGGAACAUAUAG-----AUACACAUACA-----UCAUAUAUUGCUGGAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC .((((((((((....))))......-----....((((..(-----((((.....((((((....)))))).(((....)))))))).))))......))))))................ ( -25.60) >DroSim_CAF1 98578 110 - 1 AGAAGUCGCUCACAGGAACAUAUAG-----AUACACACACA-----UCAUAUAUUGCUGGAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC .(((((((((..(((....((((((-----((........)-----)).))))).((((((....)))))))))..)))...(((((...)))))...))))))................ ( -23.40) >DroEre_CAF1 77350 120 - 1 AGAAAUCGCUCACAGGAGCAUAUAUUCGUAGCACACAUAAAUGUACAUGUACAUGGCUGAAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC .((((((((((....))))((((..((((.((((......(((((....)))))(((((........))))).....)))).))))..))))......))))))................ ( -32.90) >DroYak_CAF1 90917 101 - 1 AGAAGUCGCUCACAGGAACAUAUAU-------------------AGUCGUACAUGGCUGGAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCAAUUUCACUACAUCUACGACCC .........................-------------------.((((((...(((((((....)))))))(((....)))(((((...))))).................)))))).. ( -22.70) >consensus AGAAGUCGCUCACAGGAACAUAUAG______CACACAUACA_____UCAUACAUUGCUGGAGAACUCCGGCCUGCAAGUGCAAUGAUAAUGUCAUACCGAUUUCACUACAUCUCCAACCC .((((((((((....))))...........................((((.....((((((....)))))).(((....)))))))............))))))................ (-19.26 = -19.54 + 0.28)

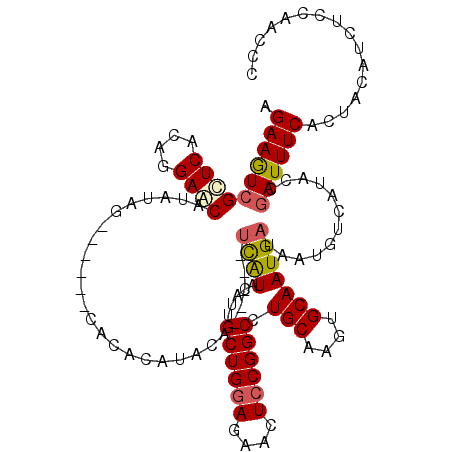
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:38:02 2006