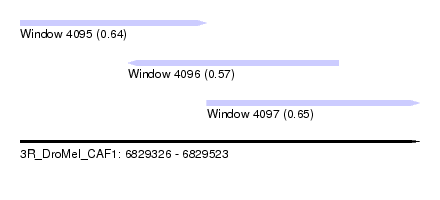
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,829,326 – 6,829,523 |
| Length | 197 |
| Max. P | 0.651176 |

| Location | 6,829,326 – 6,829,418 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6829326 92 + 27905053 ---------------------------CGACCACUGCCAAAAUUGGCGGUCAAUUAAAACGGCGGCCUGAUUGAAAUUCAUUUUCGGUCGAAAACCAAAUGGGCGU-UGAAAAAUAUGCU ---------------------------.((((...((((....))))))))........(((((.((.((((((((.....))))))))(.....)....)).)))-))........... ( -25.60) >DroSec_CAF1 88411 92 + 1 ---------------------------CGACCACUGCCAAAAUUGGUGGUCAAUUAAAACGGCGGCCUGACUGAAAUUCAUUUUAGGUCGAAAACCAACUGGGAGU-UGAAAAAUAUGCU ---------------------------.(((((((.........))))))).........((((((((((.((.....))..)))))))......((((.....))-))........))) ( -24.30) >DroSim_CAF1 94685 119 + 1 NNNNNNNNNNNNGGUGGAUUCAACUUUCGACCACUGCCAAAAUUGGCGGUCAAUUAAAACGGCGGCCUGAUUGAAAUUCAUUUUAGGUCGAAAACCAACUGGGCGU-UGAAAAAUAUGCU ..................((((((((((((((.((((((....))))))(((((((...........)))))))...........)))))))).((....))..))-))))......... ( -30.80) >DroEre_CAF1 73376 92 + 1 ---------------------------CGACCACUGCCGAAAUUGGUGGUCAAUUAAAACGGUGGCCUGAUUGAAAUUCAUUUUAGGUCGAAAACCAACUGGGCGU-UGAAAAAUAUGCU ---------------------------.(((((((.........))))))).........((((((((((.((.....))..)))))))....))).....(((((-........))))) ( -25.00) >DroYak_CAF1 86736 92 + 1 ---------------------------CGACCACUGCCGAAAUUGGCGGUCAAUUAAAACGGUGGCCUGAUUGAAAUUCAUUUUAGGGCGAAAACCAACUGGGCGU-UGAAAAAUAUGCU ---------------------------...(((((((((....)))))))..........((((.(((((.((.....))..))))).)....)))....))((((-........)))). ( -22.30) >DroAna_CAF1 44662 93 + 1 ---------------------------UGGCUACUGUCGAAAUUGGCGGUCAAUUAAAAUGGCGGCCUGAUUGAAAUUUCAUUUGUACGGAAUGUGGACGGGGCGUAUGAGUAAUAUGCU ---------------------------......(((((..(((((((.((((.......)))).))).))))...(((((........)))))...)))))(((((((.....))))))) ( -24.60) >consensus ___________________________CGACCACUGCCAAAAUUGGCGGUCAAUUAAAACGGCGGCCUGAUUGAAAUUCAUUUUAGGUCGAAAACCAACUGGGCGU_UGAAAAAUAUGCU ...........................(((((.((((((....))))))(((((((...........)))))))...........)))))...........(((((.(.....).))))) (-16.49 = -16.52 + 0.03)

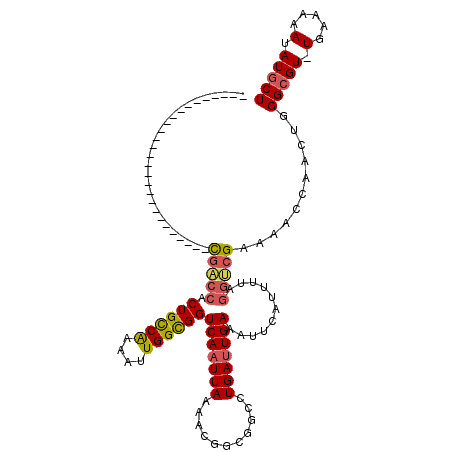
| Location | 6,829,379 – 6,829,483 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.54 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6829379 104 - 27905053 UGGUUGCCAAAGAAAAAUGGCAAA---ACAAAA-AAUGAAAAGAACAGGAGCCAGCUUCAAAAGCAGUAAGCAUAUUUUUCAACGCCCAUUUGGUUUUCGACCGAAAA (((((((((........)))))).---......-..(((((((....((((....))))....((.....))...)))))))....)))(((((((...))))))).. ( -22.80) >DroSec_CAF1 88464 99 - 1 UGGUUGCCAAAGAAAAAUGGCAAACAAACAAAAAAAA---------AUGAGCCAGCUUCAAAAGCAGUAAGCAUAUUUUUCAACUCCCAGUUGGUUUUCGACCUAAAA .((((((((........))))....((((....((((---------(((.((..(((.(....).)))..)).)))))))((((.....))))))))..))))..... ( -18.20) >DroSim_CAF1 94765 108 - 1 UGGUUGCCAAAGAAAAAUGGCAAACAAACAAAAAAAAAAAAAGAAAAGGGGCCAGCUUUAAAAGCAGUAAGCAUAUUUUUCAACGCCCAGUUGGUUUUCGACCUAAAA .((((((((........)))).................((((..((..((((..(((.....))).(....)............))))..))..)))).))))..... ( -18.90) >DroEre_CAF1 73429 102 - 1 UGGUUGCCAAAGAAAAAUUGGGAACAA------AAAUAAAAAGAACAGGGGCCAGCUUCAAAAGCAGUAAGCAUAUUUUUCAACGCCCAGUUGGUUUUCGACCUAAAA .(((((...((((..(((((((...((------(((((.........((((....))))....((.....)).))))))).....)))))))..)))))))))..... ( -22.70) >DroYak_CAF1 86789 100 - 1 UGGUUGCCAAAGAAAAAUG--GCAAAA------AAAAAAACAGAGCAGGGGUCAGCUUCAAAAGCAGUAAGCAUAUUUUUCAACGCCCAGUUGGUUUUCGCCCUAAAA ...((((((........))--))))..------..............(((((..(((((.......).)))).......(((((.....))))).....))))).... ( -20.10) >consensus UGGUUGCCAAAGAAAAAUGGCAAACAAACAAAAAAAAAAAAAGAACAGGGGCCAGCUUCAAAAGCAGUAAGCAUAUUUUUCAACGCCCAGUUGGUUUUCGACCUAAAA .(((((.....((((((((............................((((....))))....((.....)).)))))))).....((....))....)))))..... (-13.30 = -13.54 + 0.24)

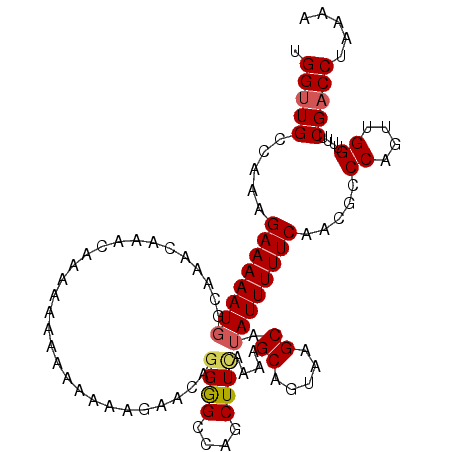
| Location | 6,829,418 – 6,829,523 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.97 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.17 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6829418 105 + 27905053 UACUGCUUUUGAAGCUGGCUCCUGUUCUUUUCAUU-UUUUGU----UUUGCCAUUU----------UUCUUUGGCAACCACAACAAAGUGUUAGAGGGUCAGGUUAAUUGAUACACAGAC ..(((..((..((.(((((((....(((...((((-((((((----.((((((...----------.....))))))..)))).))))))..)))))))))).))....))....))).. ( -25.80) >DroSec_CAF1 88503 100 + 1 UACUGCUUUUGAAGCUGGCUCAU---------UUUUUUUUGU-UUGUUUGCCAUUU----------UUCUUUGGCAACCACAACAAAGUGUUAGAAGGGCAGGUUAAUUGAUACACAGAC ..(((...(..(((((.((((.(---------(((.((((((-(((.((((((...----------.....)))))).)).)))))))....)))))))).))))..)..)....))).. ( -24.10) >DroSim_CAF1 94804 109 + 1 UACUGCUUUUAAAGCUGGCCCCUUUUCUUUUUUUUUUUUUGU-UUGUUUGCCAUUU----------UUCUUUGGCAACCACAACAAAGUGUUAGAGGGGCAGGUUAAUUGAUACACAGAC ..(((...((((((((.((((((((...........((((((-(((.((((((...----------.....)))))).)).)))))))....)))))))).))))..))))....))).. ( -30.96) >DroEre_CAF1 73468 103 + 1 UACUGCUUUUGAAGCUGGCCCCUGUUCUUUUUAUUU-------UUGUUCCCAAUUU----------UUCUUUGGCAACCACAACAAAGUGUUAGAGGGGCAGGUUAAUUGGUACACAGAC ...((((..(..((((.((((((.............-------..(((.((((...----------....)))).)))(((......)))....)))))).)))).)..))))....... ( -23.50) >DroYak_CAF1 86828 101 + 1 UACUGCUUUUGAAGCUGACCCCUGCUCUGUUUUUUU-------UUUUGC--CAUUU----------UUCUUUGGCAACCACAACAAAGUGUUAGAGGGGCAGGUUAAUUGAUACACAGAC ..(((..((..((.(((.((((((((.((((.....-------..((((--((...----------.....))))))....)))).))).....)))))))).))....))....))).. ( -24.10) >DroAna_CAF1 44755 120 + 1 UAUUGCUUUCAGUUUUUUUCCCUUUUUUUUCUUUCUGUGAUUUUUUUUACCCGUUUUUGUGUUUUAUUUUCUGGCAACCACAACAAAGUGUUAGAGGGACAGGUUAAUUGAUACGCAGAC ..((((..((((((..(((((((((.....((((.((((........(((........)))...........(....)))))..))))....)))))))..))..))))))...)))).. ( -20.70) >consensus UACUGCUUUUGAAGCUGGCCCCUGUUCUUUUUUUUUUUUUGU_UUGUUUGCCAUUU__________UUCUUUGGCAACCACAACAAAGUGUUAGAGGGGCAGGUUAAUUGAUACACAGAC ..(((...((((((((.((((((.................................................(....)(((......)))....)))))).))))..))))....))).. (-13.52 = -13.17 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:59 2006