
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,828,680 – 6,828,838 |
| Length | 158 |
| Max. P | 0.699111 |

| Location | 6,828,680 – 6,828,799 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -29.81 |
| Energy contribution | -29.87 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
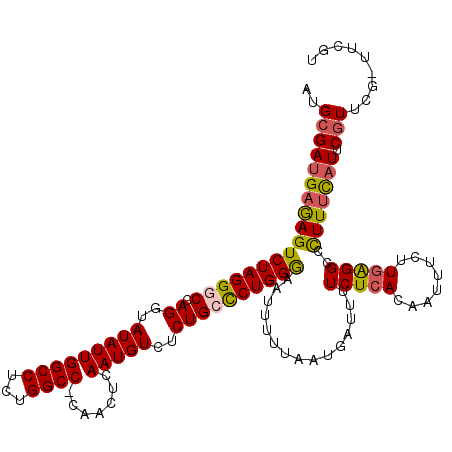
>3R_DroMel_CAF1 6828680 119 - 27905053 AAAUCAUUAAAAAUUUCCAGGGCAGAGACAUUGAGUUG-GGCCAGAGGCCAAUAUACCUGGACCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGCCAG ....................((((((.(.((..(.(((-((((...)))).....((((((..(((((..((((........))))..))))))).))))))).)..)).).)))))).. ( -33.70) >DroSec_CAF1 87789 119 - 1 AAAUCAUUAAAAAUUCCCAGAGCAGAGACAUUGAGUUG-GGCCAGAGGCCAAUAUACCUGGGCCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGCCAG ...................(.(((((.(.((..(.(((-((((...)))).....((((.(((.((((..((((........))))..))))))).))))))).)..)).).)))))).. ( -35.20) >DroSim_CAF1 93641 119 - 1 AAAUCAUUAAAAAUUCCCAGGGCAGAGACAUUGAGUUG-GGCCAGAGGCCAAUAUACCUGGCCCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGCCAG ....................((((((.(.((..(...(-((((((.(.........))))))))..((((....(((...(((((....))))))))))))...)..)).).)))))).. ( -35.50) >DroEre_CAF1 72711 119 - 1 AAAUCAUUAAAAAUUCCCAGGGCAGAGACAUUGAGUUG-GGCCAGAGGCCAAUAUACCUGGCCCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGGCAG .......((((.((((....((((..((....((((((-((((((.(.........))))))))..)))))...))...)))))))).))))(((((.(((.....)))...)).))).. ( -33.00) >DroYak_CAF1 86082 119 - 1 AAAUCAUUAAAAAUUCCCAGGGCAGAGACAUUGAGUUG-GGCCAGAGGCCAAUAUACCUGGCCCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGCCAG ....................((((((.(.((..(...(-((((((.(.........))))))))..((((....(((...(((((....))))))))))))...)..)).).)))))).. ( -35.50) >DroAna_CAF1 44046 120 - 1 AAAUCAUUAAAAAUUCCCAGGGCAGAGACAUUGAGUUGGGGCCAGAGGCCAAUAUACCUGGCCCUAGACUUUCAUCGCAUGCUGAGUAUUUAGUCGAGGUCAAAUUGAUAUGUCUGCCAG ....................((((((.(.((..(..(((((((((.(.........))))))))))((((....(((...(((((....))))))))))))...)..)).).)))))).. ( -36.50) >consensus AAAUCAUUAAAAAUUCCCAGGGCAGAGACAUUGAGUUG_GGCCAGAGGCCAAUAUACCUGGCCCUAGACUCUCAUCGCAUGCUGAGUAUUUAGCCGAGGUCAAAUUGAUAUGUCUGCCAG ....................((((((.(.((..(.(((.((((...)))).....((((((..(((((..((((........))))..))))))).))))))).)..)).).)))))).. (-29.81 = -29.87 + 0.06)



| Location | 6,828,720 – 6,828,838 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -27.49 |
| Energy contribution | -27.66 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6828720 118 + 27905053 AUGCGAUGAGAGUCUAGGUCCAGGUAUAUUGGCCUCUGGCC-CAACUCAAUGUCUCUGCCCUGGAAAUUUUUAAUGAUUUUCUCACAAUUUCUUAAGGCCCUUUCAUUCGUUCG-UUCGU ..((((((((((((((((..(((..(((((((((...))))-......)))))..))).))))))...((((((.((..((.....))..))))))))..)))))).))))...-..... ( -26.50) >DroSec_CAF1 87829 118 + 1 AUGCGAUGAGAGUCUAGGCCCAGGUAUAUUGGCCUCUGGCC-CAACUCAAUGUCUCUGCUCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGAGGCCCUUUCAUUCGUUCG-UUCGU ..((((((((((....((((((((...(((((((...))).-...................((((((..((....))..))))))))))..)))).)))))))))).))))...-..... ( -30.10) >DroSim_CAF1 93681 118 + 1 AUGCGAUGAGAGUCUAGGGCCAGGUAUAUUGGCCUCUGGCC-CAACUCAAUGUCUCUGCCCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGAGGCCCUUUCAUUCGUUCG-UUCGU ..((((((((((....(((((((((......)))..)))))-)........(((((.....((((((..((....))..)))))).........))))).)))))).))))...-..... ( -33.54) >DroEre_CAF1 72751 118 + 1 AUGCGAUGAGAGUCUAGGGCCAGGUAUAUUGGCCUCUGGCC-CAACUCAAUGUCUCUGCCCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGAGGCCCUUUCAUUCGUUCA-UUCGU ..((((((((((....(((((((((......)))..)))))-)........(((((.....((((((..((....))..)))))).........))))).)))))).))))...-..... ( -33.54) >DroYak_CAF1 86122 118 + 1 AUGCGAUGAGAGUCUAGGGCCAGGUAUAUUGGCCUCUGGCC-CAACUCAAUGUCUCUGCCCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGAGGUCCUUUCAUUCGUUGG-UUCGU ..((((((((((((((((((.((..(((((((((...))))-......)))))..))))))))))...............(((((........)))))..)))))).))))...-..... ( -33.50) >DroAna_CAF1 44086 120 + 1 AUGCGAUGAAAGUCUAGGGCCAGGUAUAUUGGCCUCUGGCCCCAACUCAAUGUCUCUGCCCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGGGGCCUUUCUUUCCAUUCGAAUCGU ...(((.(((((....(((((((.....)))))))..(((((((((.....))........((((((..((....))..))))))........)))))))...)))))...)))...... ( -35.10) >consensus AUGCGAUGAGAGUCUAGGGCCAGGUAUAUUGGCCUCUGGCC_CAACUCAAUGUCUCUGCCCUGGGAAUUUUUAAUGAUUUUCUCACAAUUUCUUGAGGCCCUUUCAUUCGUUCG_UUCGU ..((((((((((((((((((.((..(((((((((...)))).......)))))..))))))))))...............(((((........)))))..))))))).)))......... (-27.49 = -27.66 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:57 2006