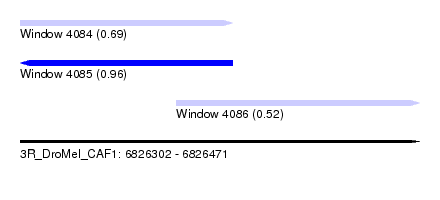
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,826,302 – 6,826,471 |
| Length | 169 |
| Max. P | 0.959558 |

| Location | 6,826,302 – 6,826,392 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -13.36 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6826302 90 + 27905053 UGUAAGCAGCCGGAAAAAAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUGAGCGGCCGAAGCACAA-------AAAAAAAAAUCAAAGAAGA-A (((...(.(((((((((((.(((((......)))))...))))))))..((....))))).)..)))...-------...................-. ( -19.30) >DroSec_CAF1 85396 86 + 1 UGUAAGCAGCCGGAAA-AAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUCAGCGGCCCAAGCACAA-------AA---AAAAUCAAAGAAGA-A .....((.((((((((-((((((((......)))))...))))))))..((....)))))....))....-------..---..............-. ( -18.90) >DroSim_CAF1 91311 90 + 1 UGUAAGCAGCCGGAAAAAAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUCAGCGGCCCAAGCACAA-------AAAAAAAAAUCAAAGAAGA-A .....((.(((((((((((.(((((......)))))...))))))))..((....)))))....))....-------...................-. ( -19.00) >DroEre_CAF1 70317 96 + 1 UGUAAGCAGCCGGAAAAAAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUCAGCGGGCCGG-CAAAAAAAAAAAGAAAAAAAAUCAAAGAAGA-A ........(((((((((((.(((((......)))))...))))))))..((((....)))).))-)..............................-. ( -22.50) >DroYak_CAF1 83638 87 + 1 UGUAAGCAGCCGGAAAAGAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUCAGCGGCCCAGGCAACA----------AAAAAAUCAAAGAAGA-A ........(((((((((((.(((((......)))))...))))))))..(((.....)))...)))....----------................-. ( -21.70) >DroAna_CAF1 41909 86 + 1 UGUAAGCAACCGA-GAAAAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUGAGCAGCCAAA-CAAAA----------AAAAAUUGAAAGAAGAAA (((..((....((-(((((.(((((......)))))...)))))))..(((....)))))...)-))...----------.................. ( -12.10) >consensus UGUAAGCAGCCGGAAAAAAAACACAUUAAAUUGUGUAAAUUUUUUCCAUGCCUCAGCGGCCCAAGCAAAA_______AA_AAAAAAUCAAAGAAGA_A ........(((((((((((.(((((......)))))...))))))))..((....)))))...................................... (-13.36 = -14.08 + 0.72)

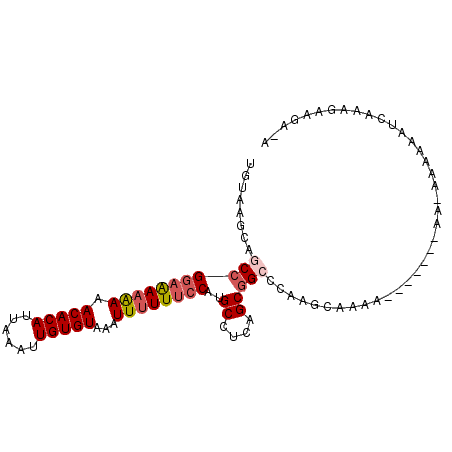

| Location | 6,826,302 – 6,826,392 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6826302 90 - 27905053 U-UCUUCUUUGAUUUUUUUUU-------UUGUGCUUCGGCCGCUCAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUUUUUUCCGGCUGCUUACA .-...................-------.((((...(((((((....))..((((((((...(((((......))))).)))))))))))))..)))) ( -23.90) >DroSec_CAF1 85396 86 - 1 U-UCUUCUUUGAUUUU---UU-------UUGUGCUUGGGCCGCUGAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUU-UUUCCGGCUGCUUACA .-..............---..-------....((...((((((....))..((((((((...(((((......))))))))-)))))))))))..... ( -19.90) >DroSim_CAF1 91311 90 - 1 U-UCUUCUUUGAUUUUUUUUU-------UUGUGCUUGGGCCGCUGAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUUUUUUCCGGCUGCUUACA .-...................-------....((...((((((....))..((((((((...(((((......))))).))))))))))))))..... ( -21.40) >DroEre_CAF1 70317 96 - 1 U-UCUUCUUUGAUUUUUUUUCUUUUUUUUUUUG-CCGGCCCGCUGAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUUUUUUCCGGCUGCUUACA .-..............................(-((.(((......)))..((((((((...(((((......))))).)))))))))))........ ( -22.00) >DroYak_CAF1 83638 87 - 1 U-UCUUCUUUGAUUUUUU----------UGUUGCCUGGGCCGCUGAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUCUUUUCCGGCUGCUUACA .-................----------.((.(((...(((.....)))..((((((.....(((((......)))))...))))))))).))..... ( -20.80) >DroAna_CAF1 41909 86 - 1 UUUCUUCUUUCAAUUUUU----------UUUUG-UUUGGCUGCUCAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUUUUC-UCGGUUGCUUACA ...........(((((((----------(((((-(((((....))))))).)))))))))).(((((......)))))......-............. ( -12.20) >consensus U_UCUUCUUUGAUUUUUU_UU_______UUGUGCUUGGGCCGCUGAGGCAUGGAAAAAAUUUACACAAUUUAAUGUGUUUUUUUUCCGGCUGCUUACA .....................................((((((....))..((((((((...(((((......))))).))))))))))))....... (-17.17 = -17.62 + 0.45)



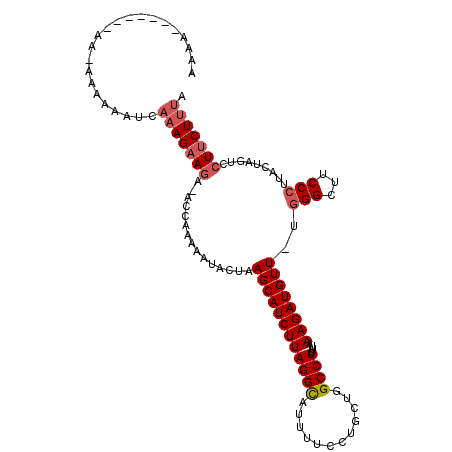
| Location | 6,826,368 – 6,826,471 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -15.54 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6826368 103 + 27905053 ACAA-------AAAAAAAAAUCAAAGAAGA-ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCUGCUGGCCUUUUAAAGAUGUU-UGGGCUUCCCUCACUAGUCCUUCUUCG ....-------..............(((((-(...........(((((((((((((............))))....))))))))-)(((((.........))))))))))). ( -23.00) >DroSec_CAF1 85461 100 + 1 ACAA-------AA---AAAAUCAAAGAAGA-ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCUGCUGGCCUUUUAAAGAUGUU-CGGGCUUCCCUUACUAGUCCUUCUUUG ....-------..---.....((((((((.-((...........((((((((((((............))))....))))))))-.(((...)))......)).)))))))) ( -21.90) >DroSim_CAF1 91377 104 + 1 ACAA-------AAAAAAAAAUCAAAGAAGA-ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCUGCUGGCCUUUUAAAGAUGUUUUGGGCUUCCCUCACUAGUCCUUCUUUG ....-------..........((((((((.-((..........(((((((((((((............))))....))))))))).(((...)))......)).)))))))) ( -22.40) >DroEre_CAF1 70382 110 + 1 AAAAAAAAAAAGAAAAAAAAUCAAAGAAGA-ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCUGCUGGCCUUUUAAAGAUGUU-UGGGCUUCCCUUACUAGUCCUCCUUUA .........................((((.-............(((((((((((((............))))....))))))))-)(((((.........)))))..)))). ( -18.50) >DroYak_CAF1 83704 100 + 1 AACA----------AAAAAAUCAAAGAAGA-ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCAGCUGGCCUUUUAAAGAUGUU-UGGGCUUCCCUUACUAGUCCUUCUUUA ....----------........(((((((.-..........(((((((((((((((............))))....))))))))-)))(((.........))).))))))). ( -22.00) >DroAna_CAF1 41973 96 + 1 AAAA----------AAAAAUUGAAAGAAGAAACCAAAAAUACUAAGCAUCUUAGGUAUUUCUUUGCCGGCCUUUUAAAGAUGUU-CAGGCUUCCCGUACU-----UUCUAUA ....----------.......(((((...............((.((((((((.((((......)))).........))))))))-.))((.....)).))-----))).... ( -14.70) >consensus AAAA_______AA_AAAAAAUCAAAGAAGA_ACCAAAAAUACUAAGCAUCUUAGGCAUUUUCCUGCUGGCCUUUUAAAGAUGUU_UGGGCUUCCCUUACUAGUCCUUCUUUA ......................(((((((...............((((((((((((............))))....))))))))..(((...))).........))))))). (-15.54 = -16.23 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:49 2006