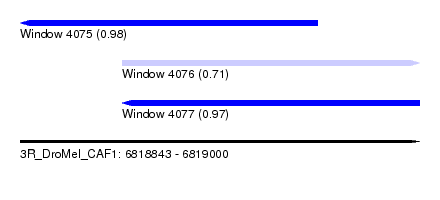
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,818,843 – 6,819,000 |
| Length | 157 |
| Max. P | 0.976018 |

| Location | 6,818,843 – 6,818,960 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6818843 117 - 27905053 AAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUAGAAUUUGACGAGGCUUAACUUU ((((((((((......((((((((....(((..(((((....)))))..))).---))))))))...((((......))))))))))))))................(((......))). ( -22.80) >DroSim_CAF1 83868 117 - 1 AAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUAGAAUUUGACGAGGCUUAACUUU ((((((((((......((((((((....(((..(((((....)))))..))).---))))))))...((((......))))))))))))))................(((......))). ( -22.80) >DroEre_CAF1 62875 117 - 1 AAUUCAUUUUAAUAUUUACAAACAUUUUUUUCCCGGCUCCUUAGCCACUCGAC---UGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUACAAUUUGACGAGGCUUAACUUU ((((((((((......((((((((.((.......((((....))))....)).---))))))))...((((......))))))))))))))................(((......))). ( -18.20) >DroYak_CAF1 76123 117 - 1 AAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUACAAUUUGACGAGGCUUAACUUU ((((((((((......((((((((....(((..(((((....)))))..))).---))))))))...((((......))))))))))))))................(((......))). ( -22.80) >DroAna_CAF1 34498 120 - 1 AAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCAAGCUCCUUGGCUGCGCGACGGUUGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUAGAAUUUGACGCGGCUCGACUUU ((((((((((....................((((((...)))))).....(((((((((.(((.....))).))))))))))))))))))).......(((.((((......)))).))) ( -26.20) >consensus AAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC___UGUUUGUACAAAACGCAUAACCGUUAAGAUGAAUUUUUAAUAGAAUUUGACGAGGCUUAACUUU ((((((((((......((((((((....(((..(((((....)))))..)))....))))))))...((((......))))))))))))))............................. (-21.64 = -21.76 + 0.12)



| Location | 6,818,883 – 6,819,000 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -19.97 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6818883 117 + 27905053 ACGGUUAUGCGUUUUGUACAAACA---GUCGAGCGGCUAAGGAGCCGGGCAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGAGCAAAAGUUUUUAAUUAAGUGAAAAGCGAA .......(((((((((((((....---.(((..(((((....))))).(((((.....)))))...........))).....)))))))))).....((((((......))))))))).. ( -26.40) >DroSim_CAF1 83908 117 + 1 ACGGUUAUGCGUUUUGUACAAACA---GUCGAGCGGCUAAGGAGCCGGGCAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGAGCAAAAGUUUUUAAUUAAGUGAAAAGCGAA .......(((((((((((((....---.(((..(((((....))))).(((((.....)))))...........))).....)))))))))).....((((((......))))))))).. ( -26.40) >DroEre_CAF1 62915 117 + 1 ACGGUUAUGCGUUUUGUACAAACA---GUCGAGUGGCUAAGGAGCCGGGAAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGAGCAAAAGUUUUUAAUUAAGUGAAAAGCGAA .......(((((((((((((....---.(((..(((((....))))).......((((((....))))))....))).....)))))))))).....((((((......))))))))).. ( -23.20) >DroYak_CAF1 76163 117 + 1 ACGGUUAUGCGUUUUGUACAAACA---GUCGAGCGGCUAAGGAGCCGGGCAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGAGCAAAAGUUUUUAAUUAAGUGAAAAGCGAA .......(((((((((((((....---.(((..(((((....))))).(((((.....)))))...........))).....)))))))))).....((((((......))))))))).. ( -26.40) >DroAna_CAF1 34538 119 + 1 ACGGUUAUGCGUUUUGUACAAACAACCGUCGCGCAGCCAAGGAGCUUGGCAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGCGG-AAAGUUUUUAAUUAAGUGAAAAACAGA ..........(((((.(((((((..((((.(((((((((((...))))))....((((((....))))))............))))).))))-...))))........))).)))))... ( -25.90) >consensus ACGGUUAUGCGUUUUGUACAAACA___GUCGAGCGGCUAAGGAGCCGGGCAAAAAAUGUUUGUAAAUAUUAAAAUGAAUUUAUGUGCAGAGCAAAAGUUUUUAAUUAAGUGAAAAGCGAA ..........((((((((((((((.........(((((....))))).........)))))((((((..........)))))))))))))))....((((((.........))))))... (-19.97 = -20.17 + 0.20)



| Location | 6,818,883 – 6,819,000 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6818883 117 - 27905053 UUCGCUUUUCACUUAAUUAAAAACUUUUGCUCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGU ...((......................(((...)))....................((((((((....(((..(((((....)))))..))).---))))))))......))........ ( -16.80) >DroSim_CAF1 83908 117 - 1 UUCGCUUUUCACUUAAUUAAAAACUUUUGCUCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGU ...((......................(((...)))....................((((((((....(((..(((((....)))))..))).---))))))))......))........ ( -16.80) >DroEre_CAF1 62915 117 - 1 UUCGCUUUUCACUUAAUUAAAAACUUUUGCUCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUUCCCGGCUCCUUAGCCACUCGAC---UGUUUGUACAAAACGCAUAACCGU ...((......................(((...)))....................((((((((.((.......((((....))))....)).---))))))))......))........ ( -12.20) >DroYak_CAF1 76163 117 - 1 UUCGCUUUUCACUUAAUUAAAAACUUUUGCUCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC---UGUUUGUACAAAACGCAUAACCGU ...((......................(((...)))....................((((((((....(((..(((((....)))))..))).---))))))))......))........ ( -16.80) >DroAna_CAF1 34538 119 - 1 UCUGUUUUUCACUUAAUUAAAAACUUU-CCGCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCAAGCUCCUUGGCUGCGCGACGGUUGUUUGUACAAAACGCAUAACCGU ...((((((.........))))))...-.(((.(((..((((.....))))...................((((((...))))))))))))((((((((.(((.....))).)))))))) ( -24.00) >consensus UUCGCUUUUCACUUAAUUAAAAACUUUUGCUCUGCACAUAAAUUCAUUUUAAUAUUUACAAACAUUUUUUGCCCGGCUCCUUAGCCGCUCGAC___UGUUUGUACAAAACGCAUAACCGU ................................(((...((((.....)))).....((((((((....(((..(((((....)))))..)))....))))))))......)))....... (-15.94 = -16.06 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:40 2006