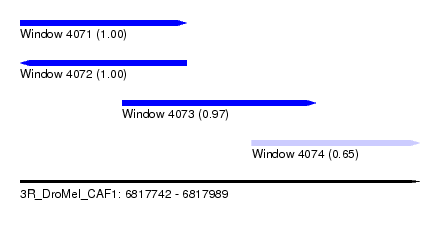
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,817,742 – 6,817,989 |
| Length | 247 |
| Max. P | 0.999921 |

| Location | 6,817,742 – 6,817,845 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -40.81 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.60 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6817742 103 + 27905053 CCUUCUCCUGGCUGCCAAA----------------CAGGCAUCUAGAAUUUUUCCCAGGCGCUGCCAGGAGAACUUCGCUUGACUGGCUGUCAUAUAAGUUUGUCAGCUUGUUUGGCUG ..((((((((((((((...----------------..))))(((.(........).)))....))))))))))....(((.(((.(((((.((........)).))))).))).))).. ( -38.00) >DroSec_CAF1 76893 106 + 1 CCUUCUCCUGGCAGCCAACCA-------------CCAGGCAUCUAUAGUUUUUCCCAGGCGCUGCCAGGAGAACUUCGCUUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUG ..(((((((((((((...((.-------------...))........((((.....)))))))))))))))))....(((.(((.(((((.((........)).))))).))).))).. ( -42.00) >DroSim_CAF1 82019 106 + 1 CCUUCUCCUGGCAGCCAACCA-------------CCAGGCAUCUAGAGUUUUUCCCAGGCGCUGCCAGGAGAACUUCGCUUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUG ..(((((((((((((...((.-------------...))........((((.....)))))))))))))))))....(((.(((.(((((.((........)).))))).))).))).. ( -42.00) >DroEre_CAF1 61784 103 + 1 CCUUCUCCUGGCAGCCAACC----------------AGCCAUCUAGAGUUUUUCCCAGGCGCUGCCAGGAGAACUUCGCUUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUG ..(((((((((((((.....----------------.(((.................))))))))))))))))....(((.(((.(((((.((........)).))))).))).))).. ( -42.13) >DroYak_CAF1 74993 119 + 1 CCUUCUCCUGGCAGCCAACCAACCAACCAGCCAAGCAGCCAUCUAGAGUUUUUCCCAGGCGCUGGCAGGAGAACUUCGCUUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUG ..((((((((.((((..............((...)).(((.................))))))).))))))))....(((.(((.(((((.((........)).))))).))).))).. ( -39.93) >consensus CCUUCUCCUGGCAGCCAACCA______________CAGGCAUCUAGAGUUUUUCCCAGGCGCUGCCAGGAGAACUUCGCUUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUG ..(((((((((((((.....................((....))...((((.....)))))))))))))))))....(((.(((.(((((.((........)).))))).))).))).. (-36.00 = -36.60 + 0.60)


| Location | 6,817,742 – 6,817,845 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.63 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -32.44 |
| Energy contribution | -33.24 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6817742 103 - 27905053 CAGCCAAACAAGCUGACAAACUUAUAUGACAGCCAGUCAAGCGAAGUUCUCCUGGCAGCGCCUGGGAAAAAUUCUAGAUGCCUG----------------UUUGGCAGCCAGGAGAAGG ((((.......))))....((((...((((.....))))....))))(((((((((.....((((((....)))))).((((..----------------...)))))))))))))... ( -37.30) >DroSec_CAF1 76893 106 - 1 CAGCCAAACAAGCUGACAAACUUGUAUGACAGCCAGUCAAGCGAAGUUCUCCUGGCAGCGCCUGGGAAAAACUAUAGAUGCCUGG-------------UGGUUGGCUGCCAGGAGAAGG ((((.......))))......((((.((((.....)))).))))..((((((((((((((((.(((..............)))..-------------.)))..))))))))))))).. ( -40.04) >DroSim_CAF1 82019 106 - 1 CAGCCAAACAAGCUGACAAACUUGUAUGACAGCCAGUCAAGCGAAGUUCUCCUGGCAGCGCCUGGGAAAAACUCUAGAUGCCUGG-------------UGGUUGGCUGCCAGGAGAAGG ((((.......))))......((((.((((.....)))).))))..(((((((((((((..(((((......)))))..(((...-------------.)))..))))))))))))).. ( -42.70) >DroEre_CAF1 61784 103 - 1 CAGCCAAACAAGCUGACAAACUUGUAUGACAGCCAGUCAAGCGAAGUUCUCCUGGCAGCGCCUGGGAAAAACUCUAGAUGGCU----------------GGUUGGCUGCCAGGAGAAGG ((((.......))))......((((.((((.....)))).))))..((((((((((((((((..(((.....)))....))).----------------.....))))))))))))).. ( -40.20) >DroYak_CAF1 74993 119 - 1 CAGCCAAACAAGCUGACAAACUUGUAUGACAGCCAGUCAAGCGAAGUUCUCCUGCCAGCGCCUGGGAAAAACUCUAGAUGGCUGCUUGGCUGGUUGGUUGGUUGGCUGCCAGGAGAAGG ((((((((((((........)))))....(((((((((((((((.....))..((((....(((((......))))).)))).))))))))))))).....)))))))........... ( -44.90) >consensus CAGCCAAACAAGCUGACAAACUUGUAUGACAGCCAGUCAAGCGAAGUUCUCCUGGCAGCGCCUGGGAAAAACUCUAGAUGCCUG______________UGGUUGGCUGCCAGGAGAAGG ((((.......))))......((((.((((.....)))).))))..(((((((((((((..(((((......)))))....(.....................)))))))))))))).. (-32.44 = -33.24 + 0.80)

| Location | 6,817,805 – 6,817,925 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.28 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -28.83 |
| Energy contribution | -28.55 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6817805 120 + 27905053 UUGACUGGCUGUCAUAUAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGG (((((((((((.((........)).)))))(((...((..(((...........)))..)).))).......))))))((((.....))))..((((..(((((....)))))..)))). ( -28.00) >DroSec_CAF1 76959 120 + 1 UUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAUGG (((((((((.((((.((((((......)))))).))((..(((...........)))..))...((((((......))))))....((((......))))...)).)))))))))..... ( -28.80) >DroSim_CAF1 82085 120 + 1 UUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGG (((.((((((..((.((((((......)))))).))((..(((...........)))..))(((((((((......))))))....((((......))))......)))))))))))).. ( -28.90) >DroEre_CAF1 61847 120 + 1 UUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGU (((.((((((..((.((((((......)))))).))((..(((...........)))..))(((((((((......))))))....((((......))))......)))))))))))).. ( -28.90) >DroYak_CAF1 75072 120 + 1 UUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGG (((.((((((..((.((((((......)))))).))((..(((...........)))..))(((((((((......))))))....((((......))))......)))))))))))).. ( -28.90) >DroAna_CAF1 33560 120 + 1 UUGACUAGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCGAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAUGU (((((((((.((((.((((((......)))))).)))).(.(((....((((((...((....))......((((((.........))))))))))))....)))))))))))))..... ( -32.00) >consensus UUGACUGGCUGUCAUACAAGUUUGUCAGCUUGUUUGGCUGCGUGUUAAACAAAAACGUGGCAACAACUUUCAAGUCAAAGUUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGG (((((((((.((((.((((((......)))))).))((..(((...........)))..))...((((((......))))))....((((......))))...)).)))))))))..... (-28.83 = -28.55 + -0.28)

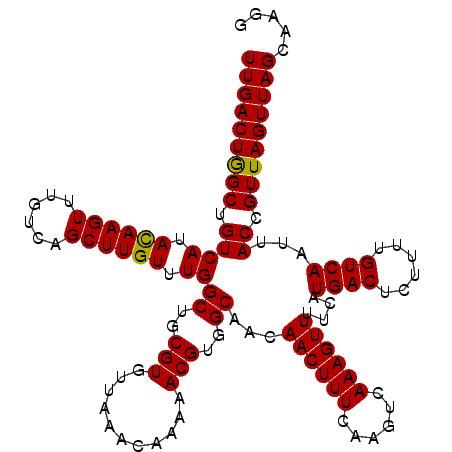
| Location | 6,817,885 – 6,817,989 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -23.31 |
| Energy contribution | -24.76 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6817885 104 + 27905053 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGGGGGUGGUGUGGGUGGCA--------------AGGGGAGAUUCCAA-GUUUUUCCUGCUUAAGACCGUGGCCAC-ACAUUA ....(((.(((((((((..(((((....)))))..))))))))).)))..(((((((--------------.((..((((.....-))))..))))))....)))(((....)-)).... ( -33.10) >DroSec_CAF1 77039 104 + 1 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAUGGGGGUGGUGUGGGUGGCA--------------AGGGGAGAUCCCAA-GUUUUCCCUGCUUAAGACCGUGGCCAC-ACAUUA ....(((.(((((..((..(((((....)))))..))..))))).)))..(((((((--------------.((((((((.....-))))))))))))....)))(((....)-)).... ( -32.60) >DroSim_CAF1 82165 104 + 1 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGGGGGUGGUGUGGGUGGCA--------------AGGGGAGAUACCAA-GUUUUCCCUGCUUAAGACCGUGGCCAC-ACAUUA ....(((.(((((((((..(((((....)))))..))))))))).)))..(((((((--------------.((((((((.....-))))))))))))....)))(((....)-)).... ( -35.60) >DroEre_CAF1 61927 118 + 1 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGUGGGUGGUGUGGGUGGUGGGGUUGGUGAGGGCAGGGGAGAUCCCAA-CUUUUACCUGCUUAAGGCCGUGGCCAC-ACAUUA ........((((.((((..(((((....)))))..)))).)))).((((((.((((.((((.((((((((...(((....)))..-)))))))).))))...))))...))))-)).... ( -40.00) >DroYak_CAF1 75152 117 + 1 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGGUGGUGGUGUGGGUGGCCGGG--GGAGGGGGGAGGUGAGAUUCCAAGGUUUUACCUGCUUAAGACCGUGGCCAC-ACAUUA ....(((.(((.(((((..(((((....)))))..))))).))).)))((.(((((((.(--(....(((.(((((((((......))))))))).)))....)).)))))))-.))... ( -44.30) >DroAna_CAF1 33640 105 + 1 UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAUGUGGGUGGUGGGGGUGCGA--------------UGUGAUGGUUUAGG-GUUUCUUCCUUCCAAGAAUGUGGCCAUUACAUUA ((..(((.((((...((..(((((....)))))..))...)))).)))..))...((--------------((((((((((....-..(((((......)))))...)))))))))))). ( -30.50) >consensus UUUUCAUGACUCUUUUGUCAAUUACCGUUAGUUAGCAAGGGGGUGGUGUGGGUGGCA______________AGGGGAGAUUCCAA_GUUUUACCUGCUUAAGACCGUGGCCAC_ACAUUA .(..(((.(((((((((..(((((....)))))..))))))))).)))..)(((((...............(((((((((......))))))))).............)))))....... (-23.31 = -24.76 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:37 2006