
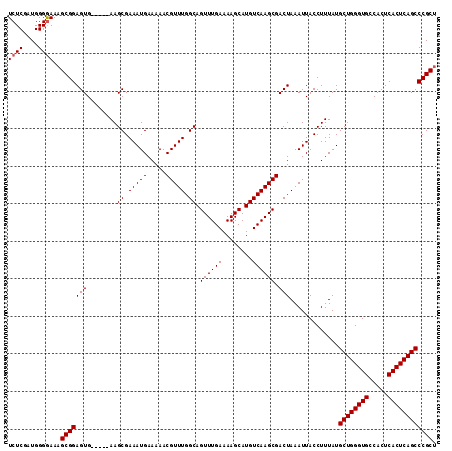
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,815,660 – 6,815,809 |
| Length | 149 |
| Max. P | 0.992013 |

| Location | 6,815,660 – 6,815,769 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.86 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -31.95 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6815660 109 + 27905053 UCUCGAUGGGGAAAGCGGAGUG-----AAGCGAAAUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCU ((((....)))).(((((.(((-----(((.(.(((......((((((((((((.....))).))))))))).....))).)))))))((((((((.....))))))))))))) ( -36.90) >DroSec_CAF1 74811 109 + 1 UCUCGAUGGGGAAAGCGGAGUG-----AAGCGAAAUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCU ((((....)))).(((((.(((-----(((.(.(((......((((((((((((.....))).))))))))).....))).)))))))((((((((.....))))))))))))) ( -36.90) >DroSim_CAF1 79921 109 + 1 UCUCGAUGGGGAAAGCGGAGUG-----AAGCGAAAUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCU ((((....)))).(((((.(((-----(((.(.(((......((((((((((((.....))).))))))))).....))).)))))))((((((((.....))))))))))))) ( -36.90) >DroEre_CAF1 59716 114 + 1 UCUCGAUGGGAAAAGCGGAGUGAAGUGAAGCGAAAUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCA (((.....)))...((((......((((((.(.(((......((((((((((((.....))).))))))))).....))).)))))))((((((((.....)))))))))))). ( -35.50) >consensus UCUCGAUGGGGAAAGCGGAGUG_____AAGCGAAAUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCU ((((....))))..(((((((........((.(((((.....))))).)).((((((........)))))).))).............((((((((.....)))))))))))). (-31.95 = -32.20 + 0.25)


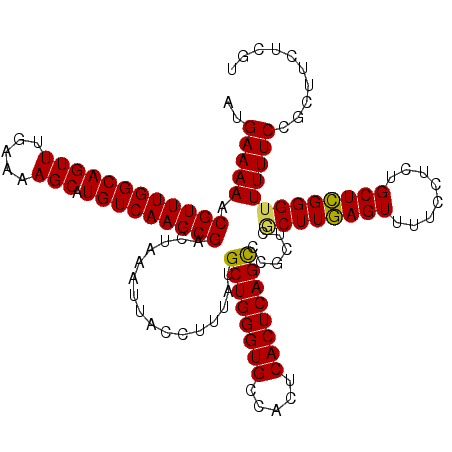
| Location | 6,815,689 – 6,815,809 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -32.16 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6815689 120 + 27905053 AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCUGGCUUGAGUUUUUCUCCGCUCGGGUUUUUCCGCUUCUCGU ..((((((((((((((((((.....))).)))))))).................((((((((.....))))))))(((..(((..(((.....))).))))))))))))).......... ( -34.90) >DroSec_CAF1 74840 120 + 1 AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCUCGCUUGAGUUUUCCUCUGCUCGGGGUUUUCCGCUUCUCUU ..(.(((((.(..(((((...(.(((((...((((((((.((((.....)))).((((((((.....))))))))....))))))))))))).).)))))..).))))).)......... ( -34.70) >DroSim_CAF1 79950 120 + 1 AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCUCGCUUGAGUUUUCCUCUGCUCGGGUUUUUCCGCUUCUCGU ..(((((((.(..(((((...(.(((((...((((((((.((((.....)))).((((((((.....))))))))....))))))))))))).).)))))..)))))))).......... ( -35.50) >DroEre_CAF1 59750 120 + 1 AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCACGCUUAAGUUUUCCUCUGCUUGGGUUUUUCCGCUUCUCUU ..(((((.((((((((((((.....))).)))))))))................((((((((.....))))))))......((((((((........))))))))))))).......... ( -31.60) >DroYak_CAF1 72886 120 + 1 AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGUCCGUUCACUUAAGUUUUCCUCUGCUUGGGUUUUUCCGCUACUCCU ..(((((.((((((((((((.....))).)))))))))................((((((((.....))))))))......((((((((........))))))))))))).......... ( -29.40) >consensus AUGAAAAACGUUUGGCAGUUUGAAAAGCAUGUCAAGCGACUAAAUUACCUUUAUGCUGGGUGCCACUCACUCAGCCCGCUCGCUUGAGUUUUCCUCUGCUCGGGUUUUUCCGCUUCUCGU ..(((((.((((((((((((.....))).)))))))))................((((((((.....))))))))......((((((((........))))))))))))).......... (-32.16 = -31.60 + -0.56)

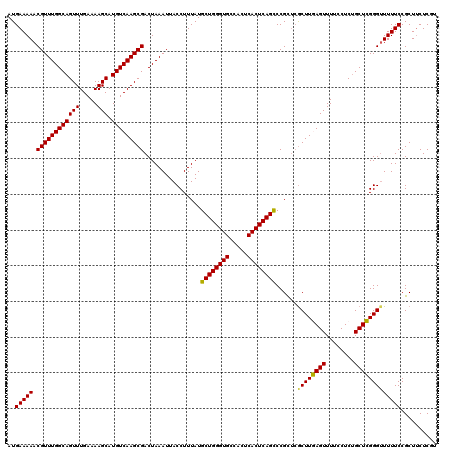
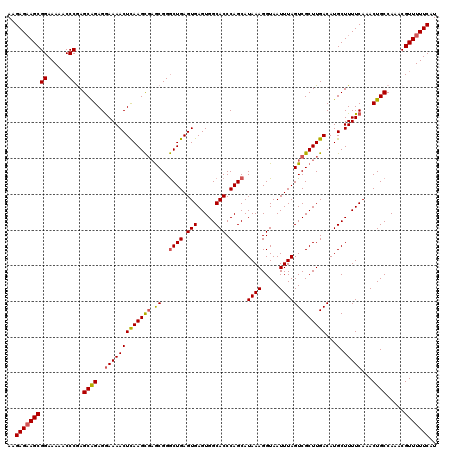
| Location | 6,815,689 – 6,815,809 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6815689 120 - 27905053 ACGAGAAGCGGAAAAACCCGAGCGGAGAAAAACUCAAGCCAGCGGGCUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..(((((((((......))(.((((......(((((.(((....))))))))(((.((((.((........)).......(((....))).)))).)))...)))))....))))))).. ( -33.00) >DroSec_CAF1 74840 120 - 1 AAGAGAAGCGGAAAACCCCGAGCAGAGGAAAACUCAAGCGAGCGGGCUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..(((((((((......))(.((((..((((((((((((((....((((.(((.....))).)))).((((.....)))).))))))))...).)))))...)))))....))))))).. ( -36.50) >DroSim_CAF1 79950 120 - 1 ACGAGAAGCGGAAAAACCCGAGCAGAGGAAAACUCAAGCGAGCGGGCUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..(((((((((......))(.((((..((((((((((((((....((((.(((.....))).)))).((((.....)))).))))))))...).)))))...)))))....))))))).. ( -36.60) >DroEre_CAF1 59750 120 - 1 AAGAGAAGCGGAAAAACCCAAGCAGAGGAAAACUUAAGCGUGCGGGCUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..(((((((((......))..((((..((((.....(((((((((((.((.(((((...(((.........))).))))).))))))).))))))))))...)))).....))))))).. ( -35.80) >DroYak_CAF1 72886 120 - 1 AGGAGUAGCGGAAAAACCCAAGCAGAGGAAAACUUAAGUGAACGGACUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..........(((((((....((((..((((((((((((((...(.(((.(((.....))).)))).((((.....)))).))))))))...).)))))...)))).....))))))).. ( -25.10) >consensus AAGAGAAGCGGAAAAACCCGAGCAGAGGAAAACUCAAGCGAGCGGGCUGAGUGAGUGGCACCCAGCAUAAAGGUAAUUUAGUCGCUUGACAUGCUUUUCAAACUGCCAAACGUUUUUCAU ..(((((((((......))..((((..(((((((((((((.((..((((.(((.....))).)))).((((.....)))))))))))))...).)))))...)))).....))))))).. (-30.62 = -30.70 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:32 2006