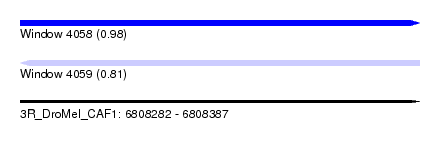
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,808,282 – 6,808,387 |
| Length | 105 |
| Max. P | 0.983366 |

| Location | 6,808,282 – 6,808,387 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -26.06 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6808282 105 + 27905053 ------------CGAAACGGACGAAAGUCGGA--AAAGCGAAAACGAGAAGCUGGGCCAAAAGAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUG ------------.....(.(((....))).).--............(.(((((((((.......((((((((((((..........))))))))))))......)).))))))).)... ( -32.62) >DroSec_CAF1 66540 105 + 1 ------------CGAAAUGGCGGCAAAUCGGA--AAAGCGAAAACGAGAAGCUGCGCCAAAAGAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUG ------------...(((((((((...(((..--..........)))...)))))((.......((((((((((((..........))))))))))))....)).)))).......... ( -32.10) >DroSim_CAF1 72111 105 + 1 ------------CGAAAGGGCCGCAAAUCGGA--AAAGCGAAAACGAGAAGCUGCGCCAAAAGAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUG ------------(....)....((((((..(.--..(((...........)))((((.......((((((((((((..........))))))))))))....)))).)..))))))... ( -32.50) >DroEre_CAF1 52274 117 + 1 AAAGCGAAAAAGCGGGAAAGCGGAAAAUCGGA--AAAGCGAAAACGGGAAGCUGCUCCAAAGGAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUC ...(((((...((((((..((((....(((..--..........)))....)))))))....((.(((((((((((..........)))))))))))))....))).....)))))... ( -35.30) >DroYak_CAF1 65160 113 + 1 ------AAAAAGCGGAAAUGCGGAAAAUCGAAAAAAAGCGAAAACGAGAAGCUGCUCCAAAGAAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUG ------.....(((((...((((....(((..............)))....)))))))......((((((((((((..........))))))))))))....))(((......)))... ( -31.54) >consensus ____________CGAAAAGGCGGAAAAUCGGA__AAAGCGAAAACGAGAAGCUGCGCCAAAAGAGUGCCAACUGGCUGAUGGGUUUGUCAGUUGGCAUCAAAGCGCAUUAGUUUGUAUG ...........................(((........)))..(((((....(((((.......((((((((((((..........))))))))))))....)))))....)))))... (-26.06 = -26.50 + 0.44)

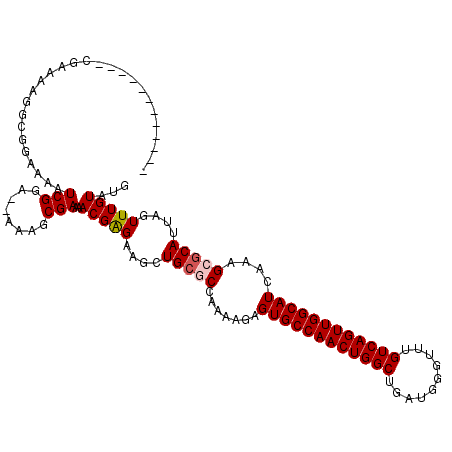
| Location | 6,808,282 – 6,808,387 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6808282 105 - 27905053 CAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUCUUUUGGCCCAGCUUCUCGUUUUCGCUUU--UCCGACUUUCGUCCGUUUCG------------ .....((((...((.(((..(((((((((((..............))))))))).))....))).))..................--...(((....))).))))..------------ ( -20.24) >DroSec_CAF1 66540 105 - 1 CAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUCUUUUGGCGCAGCUUCUCGUUUUCGCUUU--UCCGAUUUGCCGCCAUUUCG------------ ............((((((..(((((((((((..............))))))))).))....))))))..................--..(((..((....))..)))------------ ( -25.94) >DroSim_CAF1 72111 105 - 1 CAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUCUUUUGGCGCAGCUUCUCGUUUUCGCUUU--UCCGAUUUGCGGCCCUUUCG------------ ............((((((..(((((((((((..............))))))))).))....)))))).....((((..(((....--..)))...))))........------------ ( -28.54) >DroEre_CAF1 52274 117 - 1 GAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUCCUUUGGAGCAGCUUCCCGUUUUCGCUUU--UCCGAUUUUCCGCUUUCCCGCUUUUUCGCUUU .............(((....(((((((((((..............))))))))).))..((((((.(((...........))).)--))))).....)))......((......))... ( -25.24) >DroYak_CAF1 65160 113 - 1 CAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUUCUUUGGAGCAGCUUCUCGUUUUCGCUUUUUUUCGAUUUUCCGCAUUUCCGCUUUUU------ ..........(((((((((..((((((((((..............))))))))).....)..))))(((...........)))...............)))))..........------ ( -24.44) >consensus CAUACAAACUAAUGCGCUUUGAUGCCAACUGACAAACCCAUCAGCCAGUUGGCACUCUUUUGGCGCAGCUUCUCGUUUUCGCUUU__UCCGAUUUUCCGCCUUUUCG____________ ............((((((..(((((((((((..............))))))))).))....))))))...........(((........)))........................... (-19.82 = -20.26 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:22 2006