| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,804,908 – 6,805,056 |
| Length | 148 |
| Max. P | 0.988465 |

| Location | 6,804,908 – 6,805,027 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -29.33 |
| Energy contribution | -29.83 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
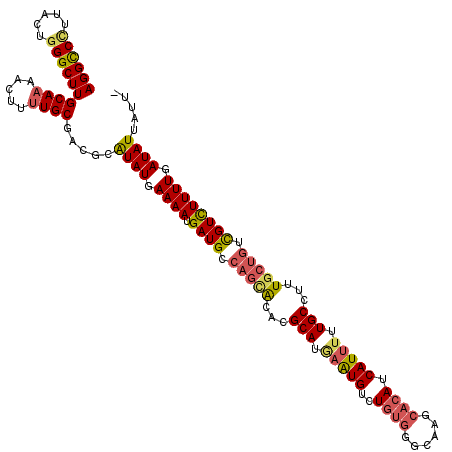
>3R_DroMel_CAF1 6804908 119 + 27905053 AGGCCCUUACUGGGCUUGCAAAACUUUUGCGACGCGUAUGAAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUU- ((((((.....))))))((((.....)))).....((((.((((.((((.(((((...(((.(((((..((((......)))).))))).)))...))))).)))))))).))))....- ( -38.10) >DroSec_CAF1 63202 119 + 1 AGGUCCUUACUGGGCUUGCAAAACUUGUGCGACGCGUAUGAAAAUGAUGCCAGCACACGCAUAAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUU- ((((((.....))))))(((.......))).....((((.((((.((((.(((((...(((.(((((..((((......)))).))))).)))...))))).)))))))).))))....- ( -34.00) >DroSim_CAF1 68745 119 + 1 AGGCCCUUACUGGGCUUGCAAAACUUUUGCGUCGCGUAUGAAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUU- ((((((.....))))))((((.....)))).....((((.((((.((((.(((((...(((.(((((..((((......)))).))))).)))...))))).)))))))).))))....- ( -38.10) >DroEre_CAF1 48924 119 + 1 AGGCCUUUGCCGGGCUUGCAAAACUUUUGCGACGCAUAUGAAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGACAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUU- .(((....)))..((((((((.....)))))).))((((.((((.((((.(((((...(((.(((((..((((......)))).))))).)))...))))).)))))))).))))....- ( -36.70) >DroYak_CAF1 61755 119 + 1 AGGCCUUUGCCGGGCUUGCAAAACUUUUGCGACGCAUAUGAAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGACAAGCACAUCAUUUUUGCCAUUCCUGCCGUCUUUUGAUAUUAUU- .(((....)))..((((((((.....)))))).))((((.((((.((((.(((.....(((.(((((..((((......)))).))))).))).....))).)))))))).))))....- ( -31.40) >DroAna_CAF1 20996 111 + 1 AGGCC-UGGCUUGGCUUGCAAAACUUUUGCGACGCAUAUGAAAACGAUGCCCCUGCACGCAUGUGUGUGU-------AGCACAUCAUU-UUGCCUUUGCCUCUGUUUUUUGAUAUUAUUU ((((.-.(((...((((((((.....)))))).)).....((((.((((...((((((((....))))))-------))..)))).))-)))))...))))................... ( -32.80) >consensus AGGCCCUUACUGGGCUUGCAAAACUUUUGCGACGCAUAUGAAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUU_ ((((((.....))))))((((.....)))).....((((.((((.((((.(((((...(((.(((((..((((......)))).))))).)))...))))).)))))))).))))..... (-29.33 = -29.83 + 0.51)


| Location | 6,804,948 – 6,805,056 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6804948 108 + 27905053 AAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUUCAUACCCUAAAAGAAAGUAUUUUCAUAUU .....((((.(((((...(((.(((((..((((......)))).))))).)))...))))).))))....((((((.(((...........)))))))))........ ( -22.90) >DroSec_CAF1 63242 107 + 1 AAAAUGAUGCCAGCACACGCAUAAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUUCAUACCCUAAAA-AAAGUAUUUUCAUAUU .....((((.(((((...(((.(((((..((((......)))).))))).)))...))))).))))...(((......)))..........-................ ( -22.40) >DroSim_CAF1 68785 107 + 1 AAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUUCAUACACUAAAA-AAAGUAUUUUCAUAUU .....((((.(((((...(((.(((((..((((......)))).))))).)))...))))).))))...(((......)))..........-................ ( -22.60) >DroEre_CAF1 48964 108 + 1 AAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGACAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUUCAUACCCUAAAAAGAGGGAUUUUCAUAUU .....((((.(((((...(((.(((((..((((......)))).))))).)))...))))).))))...(((......)))..((((......))))........... ( -28.10) >DroYak_CAF1 61795 107 + 1 AAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGACAAGCACAUCAUUUUUGCCAUUCCUGCCGUCUUUUGAUAUUAUUCAUACCCUAA-AAGAAGAAUUUUCAUAUU .....((((.(((.....(((.(((((..((((......)))).))))).))).....))).))))...(((....((((.........-.....))))..))).... ( -17.64) >consensus AAAAUGAUGCCAGCACACGCAUGAAUGUCUGUGGGCAAGCACAUCAUUUUUGCCUUUGCUGUCGUCUUUUGAUAUUAUUCAUACCCUAAAA_AAAGUAUUUUCAUAUU .....((((.(((((...(((.(((((..((((......)))).))))).)))...))))).))))...(((......)))........................... (-21.42 = -21.46 + 0.04)


| Location | 6,804,948 – 6,805,056 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.90 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6804948 108 - 27905053 AAUAUGAAAAUACUUUCUUUUAGGGUAUGAAUAAUAUCAAAAGACGACAGCAAAGGCAAAAAUGAUGUGCUUGCCCACAGACAUUCAUGCGUGUGCUGGCAUCAUUUU ...((((........(((((...(((((.....))))).)))))...(((((.(.(((..((((.((((......))))..))))..))).).)))))...))))... ( -24.70) >DroSec_CAF1 63242 107 - 1 AAUAUGAAAAUACUUU-UUUUAGGGUAUGAAUAAUAUCAAAAGACGACAGCAAAGGCAAAAAUGAUGUGCUUGCCCACAGACAUUUAUGCGUGUGCUGGCAUCAUUUU .((((....(((((((-....))))))).....)))).....((.(.(((((.(.(((.(((((.((((......))))..))))).))).).))))).).))..... ( -24.80) >DroSim_CAF1 68785 107 - 1 AAUAUGAAAAUACUUU-UUUUAGUGUAUGAAUAAUAUCAAAAGACGACAGCAAAGGCAAAAAUGAUGUGCUUGCCCACAGACAUUCAUGCGUGUGCUGGCAUCAUUUU ...((((..(((((..-....))))).....................(((((.(.(((..((((.((((......))))..))))..))).).)))))...))))... ( -22.00) >DroEre_CAF1 48964 108 - 1 AAUAUGAAAAUCCCUCUUUUUAGGGUAUGAAUAAUAUCAAAAGACGACAGCAAAGGCAAAAAUGAUGUGCUUGUCCACAGACAUUCAUGCGUGUGCUGGCAUCAUUUU ...((((.....(.((((((...(((((.....))))))))))).).(((((.(.(((..((((.((((......))))..))))..))).).)))))...))))... ( -24.30) >DroYak_CAF1 61795 107 - 1 AAUAUGAAAAUUCUUCUU-UUAGGGUAUGAAUAAUAUCAAAAGACGGCAGGAAUGGCAAAAAUGAUGUGCUUGUCCACAGACAUUCAUGCGUGUGCUGGCAUCAUUUU ...((((.......((((-(...(((((.....))))).)))))(((((.(....(((..((((.((((......))))..))))..))).).)))))...))))... ( -22.20) >consensus AAUAUGAAAAUACUUU_UUUUAGGGUAUGAAUAAUAUCAAAAGACGACAGCAAAGGCAAAAAUGAUGUGCUUGCCCACAGACAUUCAUGCGUGUGCUGGCAUCAUUUU ...((((................(((((.....))))).......(.(((((.(.(((..((((.((((......))))..))))..))).).))))).).))))... (-19.70 = -20.30 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:15 2006