
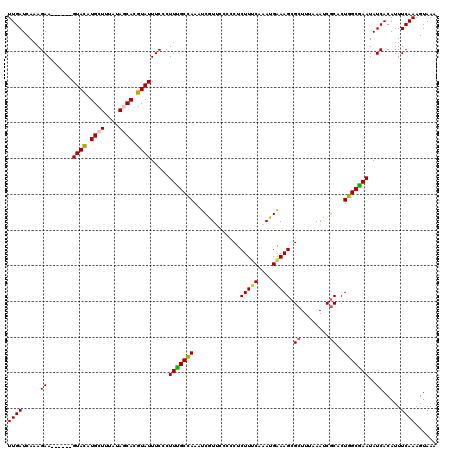
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,801,244 – 6,801,398 |
| Length | 154 |
| Max. P | 0.992005 |

| Location | 6,801,244 – 6,801,358 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.26 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6801244 114 + 27905053 UUGAUCAAAGAA------GUACAUGCUUUAUAGCACGUAUUUCUCUUUGCCAAAUCGUCCCUUCUCUUUCAAAUGGAAGCGCUUUAAAUCACACUGGCGAAUAUCACAUUUCAAAGUAAA ((((....((((------((((.((((....)))).)))))))).(((((((...........(.(((((....))))).).............))))))).........))))...... ( -23.35) >DroSec_CAF1 59584 114 + 1 UUGAUCACAGAA------GUACAUGCUUUAUAGCACGUAUUUCCCUUUGCCAAAUCAUUCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAA ((((.....(((------((((.((((....)))).)))))))..(((((((.............(((((....))))).((........))..))))))).........))))...... ( -25.50) >DroSim_CAF1 65010 114 + 1 UUGAUCAAAGAA------GUACAUGCUUUAUAGCACGUAUUUCCCUUUGCCAAAUCAUCCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAA ((((.....(((------((((.((((....)))).)))))))..(((((((.............(((((....))))).((........))..))))))).........))))...... ( -25.50) >DroEre_CAF1 45143 120 + 1 UUGAUCAAAGAAUGUUUAGUAUUUGCUGUAUAUCACGUAUUUCGCUUCGCCAAAUCGUUCCCCCUCUUACAAAUGAAAGCGCUUUAAAUCGCCCUGGCGAAUAUCACAUUUCAAAGUAAA ((((.....((.(((.((((....)))).)))))...........(((((((..(((((............)))))..(((........)))..))))))).........))))...... ( -16.60) >DroYak_CAF1 58021 120 + 1 UUGAUCAAAGAACGUUUAGUACAUGCUUUUUAUCACGUAUUUCGCUUUGCUAAAUCGUUUUCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCAAAUAUCACAUUUCAAAGUAAA .......................((((((.......((((((.(((.(((((((.(((((((............))))))).))))....)))..)))))))))........)))))).. ( -16.96) >consensus UUGAUCAAAGAA______GUACAUGCUUUAUAGCACGUAUUUCCCUUUGCCAAAUCGUUCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAA ((((.....((.......((((.((((....)))).)))).....(((((((.............(((((....))))).((........))..)))))))..)).....))))...... (-16.00 = -16.04 + 0.04)


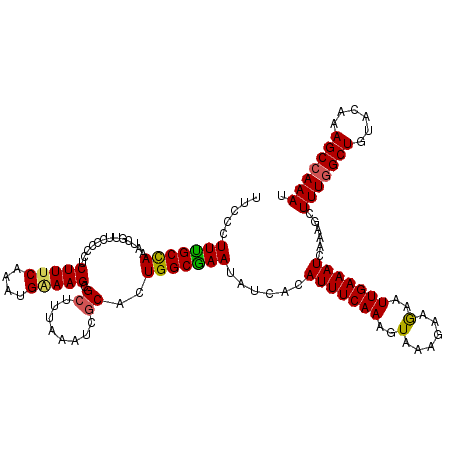
| Location | 6,801,278 – 6,801,398 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6801278 120 + 27905053 UUCUCUUUGCCAAAUCGUCCCUUCUCUUUCAAAUGGAAGCGCUUUAAAUCACACUGGCGAAUAUCACAUUUCAAAGUAAAGAAGAAUUGAAAUCAAAGCUUUUGCUGUACAAAGCCAAAU ....(((((.....(((...(((((((((.(((((((..((((............))))....)).))))).))))...)))))...)))......(((....)))...)))))...... ( -19.90) >DroSec_CAF1 59618 120 + 1 UUCCCUUUGCCAAAUCAUUCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAAGAAGAAUUGAAAUCAAAGCUUUGGCUGUACAAAGCCAAAU .....(((((((.............(((((....))))).((........))..)))))))......(((((((............)))))))......(((((((......))))))). ( -23.30) >DroSim_CAF1 65044 120 + 1 UUCCCUUUGCCAAAUCAUCCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAAGAAGAAUUGAAAUCAAAGCUUUGGCUGUACAAAGCCAAAU .....(((((((.............(((((....))))).((........))..)))))))......(((((((............)))))))......(((((((......))))))). ( -23.30) >DroEre_CAF1 45183 120 + 1 UUCGCUUCGCCAAAUCGUUCCCCCUCUUACAAAUGAAAGCGCUUUAAAUCGCCCUGGCGAAUAUCACAUUUCAAAGUAAAGAAAAAUUGAAAUCAAAGCUUUGGCUGUACAAAGCCAAAU ...(((((((((..(((((............)))))..(((........)))..)))))).......(((((((............)))))))...)))(((((((......))))))). ( -24.80) >DroYak_CAF1 58061 120 + 1 UUCGCUUUGCUAAAUCGUUUUCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCAAAUAUCACAUUUCAAAGUAAAGAAAAAUUGAAAUCAAAGCUUUGGCUGUACAAAGCCAAAU ...((((((........(((((...((((.(((((...(((........)))..(((......)))))))).))))....)))))........))))))(((((((......))))))). ( -25.49) >consensus UUCCCUUUGCCAAAUCGUUCCCCCUCUUUCAAAUGAAAGCGCUUUAAAUCGCACUGGCGAAUAUCACAUUUCAAAGUAAAGAAGAAUUGAAAUCAAAGCUUUGGCUGUACAAAGCCAAAU .....(((((((.............(((((....))))).((........))..)))))))......(((((((..(......)..)))))))......(((((((......))))))). (-20.58 = -20.34 + -0.24)

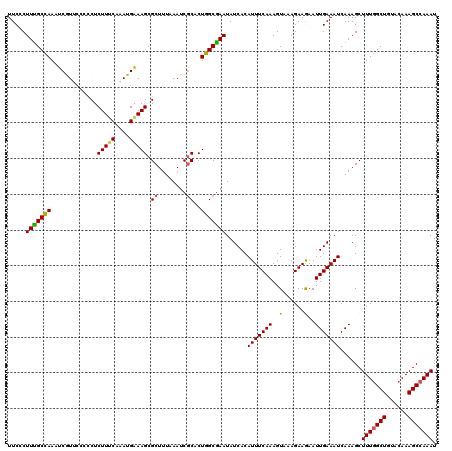
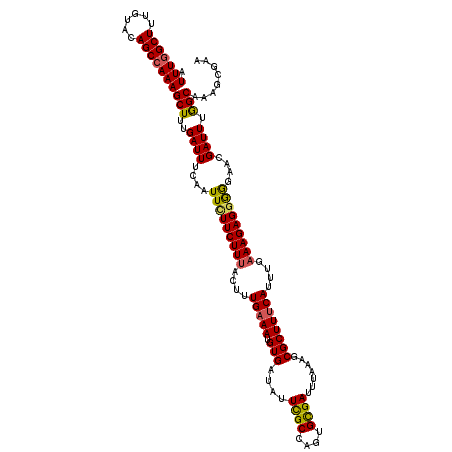
| Location | 6,801,278 – 6,801,398 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6801278 120 - 27905053 AUUUGGCUUUGUACAGCAAAAGCUUUGAUUUCAAUUCUUCUUUACUUUGAAAUGUGAUAUUCGCCAGUGUGAUUUAAAGCGCUUCCAUUUGAAAGAGAAGGGACGAUUUGGCAAAGAGAA ......((((((..(((....))).......(((.((((((((.((((.(((((.((........(((((........)))))))))))).))))..)))))).)).))))))))).... ( -24.20) >DroSec_CAF1 59618 120 - 1 AUUUGGCUUUGUACAGCCAAAGCUUUGAUUUCAAUUCUUCUUUACUUUGAAAUGUGAUAUUCGCCAGUGCGAUUUAAAGCGCUUUCAUUUGAAAGAGGGGGAAUGAUUUGGCAAAGGGAA .(((((((......))))))).(((((...(((.(((((((((....(((((.(((....((((....)))).......))))))))....)))))))))...))).....))))).... ( -32.80) >DroSim_CAF1 65044 120 - 1 AUUUGGCUUUGUACAGCCAAAGCUUUGAUUUCAAUUCUUCUUUACUUUGAAAUGUGAUAUUCGCCAGUGCGAUUUAAAGCGCUUUCAUUUGAAAGAGGGGGGAUGAUUUGGCAAAGGGAA .(((((((......))))))).(((((...(((.(((((((((....(((((.(((....((((....)))).......))))))))....)))))))))...))).....))))).... ( -32.80) >DroEre_CAF1 45183 120 - 1 AUUUGGCUUUGUACAGCCAAAGCUUUGAUUUCAAUUUUUCUUUACUUUGAAAUGUGAUAUUCGCCAGGGCGAUUUAAAGCGCUUUCAUUUGUAAGAGGGGGAACGAUUUGGCGAAGCGAA .(((((((......)))))))((((((...((..(((..((((((..(((((.(((....((((....)))).......))))))))...)).))))..)))..)).....))))))... ( -35.20) >DroYak_CAF1 58061 120 - 1 AUUUGGCUUUGUACAGCCAAAGCUUUGAUUUCAAUUUUUCUUUACUUUGAAAUGUGAUAUUUGCCAGUGCGAUUUAAAGCGCUUUCAUUUGAAAGAGGGAAAACGAUUUAGCAAAGCGAA .(((((((......)))))))((((((........((((((((.((((.(((((...........(((((........)))))..))))).))))))))))))........))))))... ( -33.11) >consensus AUUUGGCUUUGUACAGCCAAAGCUUUGAUUUCAAUUCUUCUUUACUUUGAAAUGUGAUAUUCGCCAGUGCGAUUUAAAGCGCUUUCAUUUGAAAGAGGGGGAACGAUUUGGCAAAGCGAA .(((((((......)))))))(((..((((....(((((((((....(((((.(((....((((....)))).......))))))))....)))))))))....)))).)))........ (-27.88 = -27.72 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:09 2006