
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,800,411 – 6,800,570 |
| Length | 159 |
| Max. P | 0.953179 |

| Location | 6,800,411 – 6,800,530 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.55 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -31.07 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
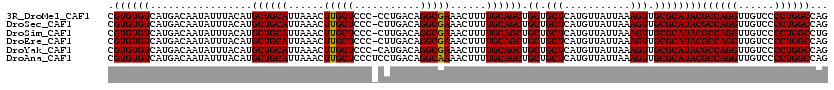
>3R_DroMel_CAF1 6800411 119 + 27905053 CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAGG-GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.(((((((((((((.(((((.((.....))-.)))))...))...)))))).)))......)))))))....... ( -34.70) >DroSec_CAF1 59029 119 + 1 CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAAG-GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.(((((((((((((.(((((.(((....))-))))))...))...)))))).)))......)))))))....... ( -34.30) >DroSim_CAF1 64164 119 + 1 CAGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAAG-GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.(((((((((((((.(((((.(((....))-))))))...))...)))))).)))......)))))))....... ( -34.30) >DroEre_CAF1 44291 119 + 1 CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAAG-GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.(((((((((((((.(((((.(((....))-))))))...))...)))))).)))......)))))))....... ( -34.30) >DroYak_CAF1 57158 119 + 1 CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAUG-GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))((..(((.((((((.......(((((....))).)))))))).))).(((((((-(.(((((.(((......))).))).....)).)))))))).)). ( -33.61) >DroAna_CAF1 16833 120 + 1 CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUUGCCUGUCAGGAGGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.(((((((((((.....(((((((.(....).)).))))).....)))))).)))......)))))))....... ( -34.70) >consensus CUGGCCAGGGGACAACCUGGCGUAUGCGCAACUUUAAUAACAUGAGCAGCAGCUGCAAAAGUUUCGCCUGUCAAG_GGGAGCAAGUUUAAUGCAGCAUGUAAAUAUUGUCAUGACACACG ...((((((......))))))...................(((((.((((((((((((((.((.(.(((.......))).).)).)))..)))))).)))......)))))))....... (-31.07 = -30.93 + -0.14)


| Location | 6,800,411 – 6,800,530 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.55 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -32.49 |
| Energy contribution | -32.35 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.03 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
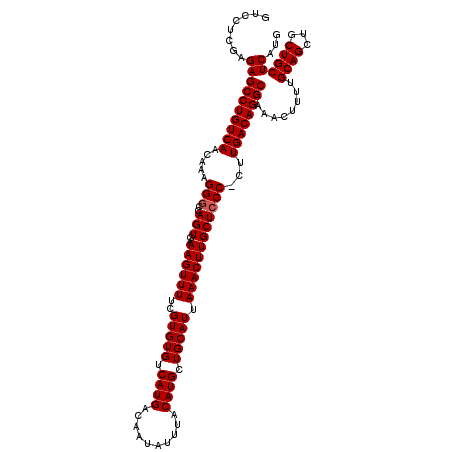
>3R_DroMel_CAF1 6800411 119 - 27905053 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CCUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG .((((((.................((((((......(((((...-.......)))))......)))))).((.(((...........))).))))))))((((((......))))))... ( -32.40) >DroSec_CAF1 59029 119 - 1 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG .((((((.................((((((......(((((...-.......)))))......)))))).((.(((...........))).))))))))((((((......))))))... ( -32.40) >DroSim_CAF1 64164 119 - 1 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCUG .((((((.................((((((......(((((...-.......)))))......)))))).((.(((...........))).))))))))((((((......))))))... ( -32.40) >DroEre_CAF1 44291 119 - 1 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG .((((((.................((((((......(((((...-.......)))))......)))))).((.(((...........))).))))))))((((((......))))))... ( -32.40) >DroYak_CAF1 57158 119 - 1 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CAUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG (((.(((((((....(((....)))..(((.......)))....-))))))).)))........((((((...((....))......))))))......((((((......))))))... ( -32.70) >DroAna_CAF1 16833 120 - 1 CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCCUCCUGACAGGCAAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG .((((((.................((((((......(((((...........)))))......)))))).((.(((...........))).))))))))((((((......))))))... ( -32.60) >consensus CGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC_CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUGUUAUUAAAGUUGCGCAUACGCCAGGUUGUCCCCUGGCCAG .((((((.................((((((......(((((...........)))))......)))))).((.(((...........))).))))))))((((((......))))))... (-32.49 = -32.35 + -0.14)


| Location | 6,800,451 – 6,800,570 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6800451 119 - 27905053 GUCCUCAAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CCUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ........((((((((((.....(((..(((..((((((..(((((.((((..........)))).))))).)))))))))..)-)))))))))).........((((...))))))... ( -32.00) >DroSec_CAF1 59069 119 - 1 GUCCUCGAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ......(((.((((((((....((((..(((..((((((..(((((.((((..........)))).))))).))))))))).))-)))))))))).........((....))..)))... ( -33.40) >DroSim_CAF1 64204 119 - 1 GUCCUCGAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ......(((.((((((((....((((..(((..((((((..(((((.((((..........)))).))))).))))))))).))-)))))))))).........((....))..)))... ( -33.40) >DroEre_CAF1 44331 119 - 1 GUCCUCGAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ......(((.((((((((....((((..(((..((((((..(((((.((((..........)))).))))).))))))))).))-)))))))))).........((....))..)))... ( -33.40) >DroYak_CAF1 57198 119 - 1 GUCCUCGAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC-CAUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ......((((((((........))(((..(..(((((((.(((.(((((((....(((....)))..(((.......)))....-))))))).))))))))))..).)))))).)))... ( -33.60) >DroAna_CAF1 16873 120 - 1 GUCCUCGUGAGCCUGUCAACAAAGGACCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCCUCCUGACAGGCAAAACUUUUGCAGCUGCUGCUCAUG .....(((((((((((((....(((....(((((.(((.(((((...))))).))).))))).....(((.......)))..)))..)))))))).........((((...))))))))) ( -33.30) >consensus GUCCUCGAGAGCCUGUCAACAAAGGGCCAGUGAAAGUUUUCGUGUGUCAUGACAAUAUUUACAUGCUGCAUUAAACUUGCUCCC_CUUGACAGGCGAAACUUUUGCAGCUGCUGCUCAUG ........((((((((((.....(((..(((..((((((..(((((.((((..........)))).))))).))))))))))))...)))))))).........((((...))))))... (-30.27 = -30.43 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:06 2006