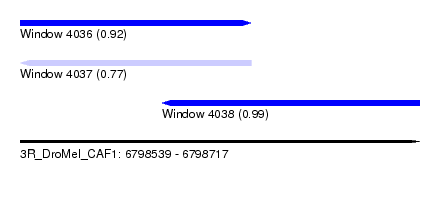
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,798,539 – 6,798,717 |
| Length | 178 |
| Max. P | 0.986856 |

| Location | 6,798,539 – 6,798,642 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.70 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -30.77 |
| Energy contribution | -31.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
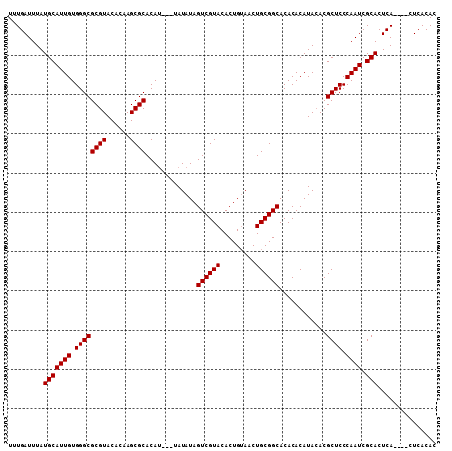
>3R_DroMel_CAF1 6798539 103 + 27905053 AUGUGAG----UGAGUGCGAUUGGGAGCGUGUAUGUGUGUGCCGCAGUUACAGUGUACGACUAUAUA--UAUGUGCGCUUGUGUACGCGCCCACAAUGCAUAAAUCAAA ...(((.----...((((.((((((.(((((((..((.((((.(((..((.(((.....))).))..--..))))))).))..))))))))).))))))))...))).. ( -33.40) >DroSec_CAF1 57145 105 + 1 GUGUGAG----UGAGUGCGAUUGGGAGCGUUUAUGUGUGUGCCGCAGUUACAGUGUACGACUAUAUAUGUAUGUGCGCUUGUGUACGCGCCCACAAUGCAUAAAUCAAA ...(((.----...(((((..((((.((((...((((.....))))..(((((((((((((.......)).))))))).)))).)))).))))...)))))...))).. ( -31.30) >DroSim_CAF1 62283 105 + 1 GUGUGAGUGAGUGAGUGCGAUUGGGAGCGUGUAUGUGUGUGCCGCAGUUACAGUGUACGACUAUAUA----UGUGCGCUUGUGUACGCGCCCACAAUGCAUAAAUCAAA .......(((....((((.((((((.(((((((..((.((((.(((..((.(((.....))).))..----))))))).))..))))))))).))))))))...))).. ( -33.80) >DroYak_CAF1 55296 101 + 1 GUGUGAG----UGAGUGCGAUUGGGAGCGUGUAUGUGUGUGCCGCAGUUCCAGUGUACGACUAUAU----AUGUGCGCUGGUGUACGCGCCCACAAUGCAUAAAUCAAA ...(((.----...((((.((((((.(((((((((((.....)))....((((((((((.......----.)))))))))).)))))))))).))))))))...))).. ( -38.40) >consensus GUGUGAG____UGAGUGCGAUUGGGAGCGUGUAUGUGUGUGCCGCAGUUACAGUGUACGACUAUAUA___AUGUGCGCUUGUGUACGCGCCCACAAUGCAUAAAUCAAA ...........(((((((.((((((.(((((((..(..((((.(((......(((((....))))).....)))))))..)..))))))))).))))))))...))).. (-30.77 = -31.02 + 0.25)

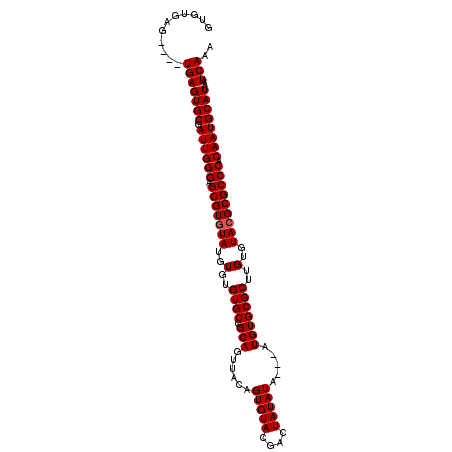
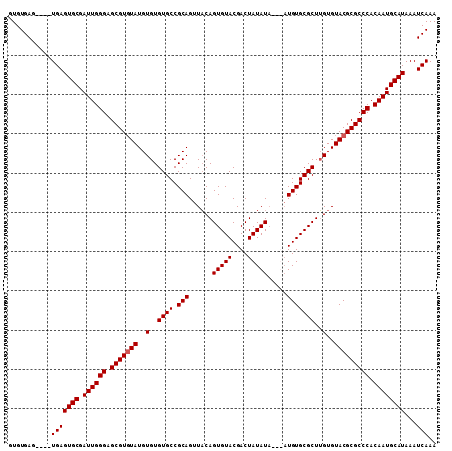
| Location | 6,798,539 – 6,798,642 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.70 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
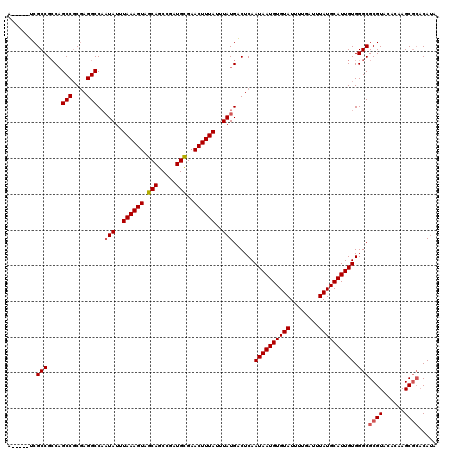
>3R_DroMel_CAF1 6798539 103 - 27905053 UUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAUA--UAUAUAGUCGUACACUGUAACUGCGGCACACACAUACACGCUCCCAAUCGCACUCA----CUCACAU ..(((....(((((((.((((((((......)))).....--......((((((.........))))))............)))).)))).)))....----.)))... ( -23.20) >DroSec_CAF1 57145 105 - 1 UUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAUACAUAUAUAGUCGUACACUGUAACUGCGGCACACACAUAAACGCUCCCAAUCGCACUCA----CUCACAC ..(((....(((((((.((((((((......)))).............((((((.........))))))............)))).)))).)))....----.)))... ( -23.20) >DroSim_CAF1 62283 105 - 1 UUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACA----UAUAUAGUCGUACACUGUAACUGCGGCACACACAUACACGCUCCCAAUCGCACUCACUCACUCACAC ..(((....(((((((.((((((((......))))...----......((((((.........))))))............)))).)))).))).....)))....... ( -23.20) >DroYak_CAF1 55296 101 - 1 UUUGAUUUAUGCAUUGUGGGCGCGUACACCAGCGCACAU----AUAUAGUCGUACACUGGAACUGCGGCACACACAUACACGCUCCCAAUCGCACUCA----CUCACAC ..(((....(((((((.((((((((......))))....----.....((((((.........))))))............)))).)))).)))....----.)))... ( -23.20) >consensus UUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAU___UAUAUAGUCGUACACUGUAACUGCGGCACACACAUACACGCUCCCAAUCGCACUCA____CUCACAC .........(((((((.((((((((......)))).............((((((.........))))))............)))).)))).)))............... (-23.10 = -23.10 + -0.00)

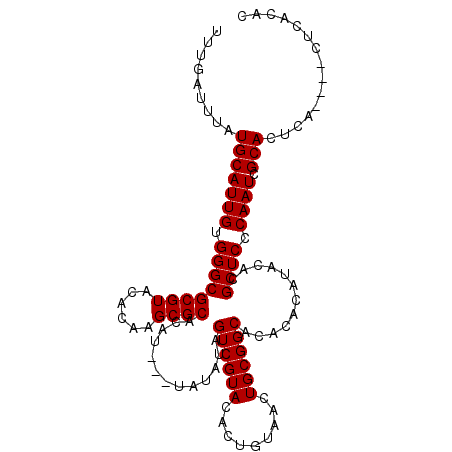
| Location | 6,798,602 – 6,798,717 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.42 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
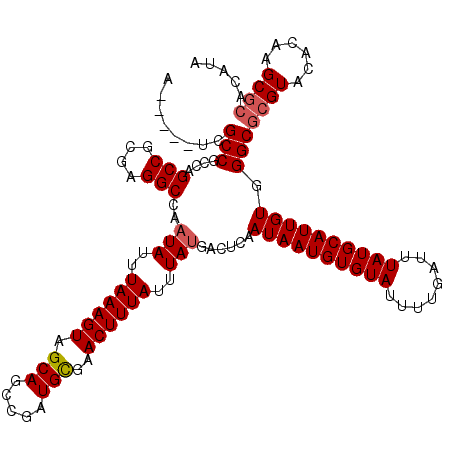
>3R_DroMel_CAF1 6798602 115 - 27905053 A-----UCGCCGCCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGGCUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAUA .-----..((((((.(((....)))..(((..((((((.(((.....)))..))))))..))))))...((((((((((........)))))))))).)))((((......))))..... ( -39.60) >DroSec_CAF1 57210 115 - 1 A-----UCGCCGCCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAUA .-----.....(((.(((....)))..(((..((((((.(((.....)))..))))))..)))......((((((((((........)))))))))).)))((((......))))..... ( -33.90) >DroSim_CAF1 62350 113 - 1 A-----UCGCCGCCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACA-- .-----.....(((.(((....)))..(((..((((((.(((.....)))..))))))..)))......((((((((((........)))))))))).)))((((......))))...-- ( -33.90) >DroEre_CAF1 42537 115 - 1 A-----UCGCCGCCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACUAAAGCGCACAUA .-----.....(((.(((....)))..(((..((((((.(((.....)))..))))))..)))......((((((((((........)))))))))).)))((((......))))..... ( -33.90) >DroYak_CAF1 55358 114 - 1 A-----UCGCCGGCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACACCAGCGCACAU- .-----..(((.((.(((....)))..(((..((((((.(((.....)))..))))))..)))........((((((((........)))))))))).)))((((......))))....- ( -34.90) >DroAna_CAF1 15089 119 - 1 GUUCCGUCGCCGCCCGCCUCAAGGCC-UUAUUUAAAGUAGCAGCCGAUGUGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACUCCAGCUACCAUA .....(((((.(((((((....))).-.((..((((((.(((.....)))..))))))..)).......((((((((((........))))))))))))))))).))............. ( -30.70) >consensus A_____UCGCCGCCAGCCGCGAGGCCAAUAUUUAAAGUAGCAGCCGAUGCGAACUUUAUUUAUGACUCAAUAAUGUGUAUUUUGAUUUAUGCAUUGUGGGCGCGUACACAAGCGCACAUA ........(((....(((....)))..(((..((((((.(((.....)))..))))))..)))......((((((((((........)))))))))).)))((((......))))..... (-31.06 = -31.42 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:37:01 2006