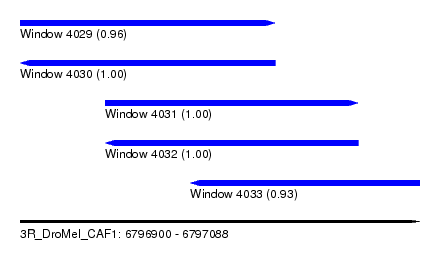
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,796,900 – 6,797,088 |
| Length | 188 |
| Max. P | 0.999920 |

| Location | 6,796,900 – 6,797,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -32.16 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6796900 120 + 27905053 UACAUGCUUACACCCCAUUGUAGUAGAGUUUUUUUUGUAAGCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUC ..((((((((((((...((((((...........))))))..))))))).)))))...........(((((((((((((..((.((...)).))..)))))))))))))........... ( -35.90) >DroSec_CAF1 55464 119 + 1 UACAUGCUUACACCCCAUUGUAGUAGAGUUU-UUUUGUAAGCGGUGUGAUGGGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUC ....(.(((((((((((((((....(((...-.)))....))))))....)))))...........(((((((((((((..((.((...)).))..))))))))))))).)))).).... ( -34.00) >DroSim_CAF1 60601 119 + 1 UACAUGCUUACACCCCAUUGUAGUAGAGUUU-UUUUGUAAGCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUC ..((((((((((((...((......))((((-......))))))))))).)))))...........(((((((((((((..((.((...)).))..)))))))))))))........... ( -36.20) >DroEre_CAF1 40772 119 + 1 UACAUGCUUACACCCCAUUGUAGUAGAUUUU-UUUUGUAAGCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUCAGUUGUAGCGGUGCCAUUUUUUAGCCAAAACCUUGACGAAAUC ..((((((((((((...((((((........-..))))))..))))))).)))))((((((.....(((...((((.((..((.((...)).))..)).))))...)))....)))))). ( -25.00) >DroYak_CAF1 53581 119 + 1 UACAUGCUUACACCCCAUUGUAGUAGAGUUU-UUUUGUAAGCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGACGAAAUC ..((((((((((((...((......))((((-......))))))))))).)))))((((((.....(((((((((((((..((.((...)).))..)))))))))))))....)))))). ( -38.40) >consensus UACAUGCUUACACCCCAUUGUAGUAGAGUUU_UUUUGUAAGCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUC ..((((((((((((...((((((...........))))))..))))))).)))))...........(((((((((((((..((.((...)).))..)))))))))))))........... (-32.16 = -32.96 + 0.80)



| Location | 6,796,900 – 6,797,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -25.03 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6796900 120 - 27905053 GAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAAAAAACUCUACUACAAUGGGGUGUAAGCAUGUA (((((......((((((((.(((....((((...))))....))).)))))))).......))))).......(((..((((((.......((((((......)))))))))))).))). ( -28.43) >DroSec_CAF1 55464 119 - 1 GAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACCCAUCACACCGCUUACAAAA-AAACUCUACUACAAUGGGGUGUAAGCAUGUA (((((......((((((((.(((....((((...))))....))).)))))))).......))))).......(((..((((((....-..((((((......)))))))))))).))). ( -28.62) >DroSim_CAF1 60601 119 - 1 GAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAA-AAACUCUACUACAAUGGGGUGUAAGCAUGUA (((((......((((((((.(((....((((...))))....))).)))))))).......))))).......(((..((((((....-..((((((......)))))))))))).))). ( -28.62) >DroEre_CAF1 40772 119 - 1 GAUUUCGUCAAGGUUUUGGCUAAAAAAUGGCACCGCUACAACUGAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAA-AAAAUCUACUACAAUGGGGUGUAAGCAUGUA (((..(((...(((((((((((.....))))...........((......)).......)))))))))).)))(((..(((((((...-....((((......)))).))))))).))). ( -17.50) >DroYak_CAF1 53581 119 - 1 GAUUUCGUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAA-AAACUCUACUACAAUGGGGUGUAAGCAUGUA (((((.(....((((((((.(((....((((...))))....))).)))))))).....).))))).......(((..((((((....-..((((((......)))))))))))).))). ( -29.30) >consensus GAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGCUUACAAAA_AAACUCUACUACAAUGGGGUGUAAGCAUGUA (((((......((((((((.(((....((((...))))....))).)))))))).......))))).......(((..(((((((.......(((((......)))))))))))).))). (-25.03 = -25.83 + 0.80)



| Location | 6,796,940 – 6,797,059 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.58 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6796940 119 + 27905053 GCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUCAAUCAGGAAAUUCUGAUUAAUUAAUCACUCAGCU-GGGCU ((.(.(((((....((((((..(.(.(((((((((((((..((.((...)).))..))))))))))))).).).))))))(((((((....))))))).....))))).).)).-..... ( -38.40) >DroSec_CAF1 55503 119 + 1 GCGGUGUGAUGGGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUCAAUCAGGAAAUUCUGAUUAAUUAAUCACUCAGCC-GGGCU .((((....(((((((((........(((((((((((((..((.((...)).))..))))))))))))).......(((.(((((((....))))))).)))))))))))))))-).... ( -41.10) >DroSim_CAF1 60640 119 + 1 GCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUCAAUCAGGAAAUUCUGAUUAAUUAAUCACUCAGCU-GGGCU ((.(.(((((....((((((..(.(.(((((((((((((..((.((...)).))..))))))))))))).).).))))))(((((((....))))))).....))))).).)).-..... ( -38.40) >DroEre_CAF1 40811 119 + 1 GCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUCAGUUGUAGCGGUGCCAUUUUUUAGCCAAAACCUUGACGAAAUCAAUCAGAAAAUUCUGAUUAAUUAAUCACUCUGCU-UGGCC ((((.(((((....(((((((.....(((...((((.((..((.((...)).))..)).))))...)))....)))))))(((((((....))))))).....))))).)))).-..... ( -32.20) >DroYak_CAF1 53620 120 + 1 GCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGACGAAAUCAAUCGGGAAAUUCUGAUUAAUUAAGCUCUCGGUUUUGGCU ((.(((.((.((..(((((((.....(((((((((((((..((.((...)).))..)))))))))))))....)))))))(((((((....)))))))......)))).))....).)). ( -35.80) >consensus GCGGUGUGAUGCGUGAUUUUGAUGGUGGUGUAGGUUAAGUUGUAGCGGUGCCAUUUUUUAGCCUACACCUUGAGGAAAUCAAUCAGGAAAUUCUGAUUAAUUAAUCACUCAGCU_GGGCU ((.(.(((((....((((((......(((((((((((((..((.((...)).))..))))))))))))).....))))))(((((((....))))))).....))))).).))....... (-34.06 = -34.58 + 0.52)



| Location | 6,796,940 – 6,797,059 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -26.36 |
| Energy contribution | -27.56 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6796940 119 - 27905053 AGCCC-AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGC .....-.(.((.((((((((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))........)))))).)).)....... ( -33.00) >DroSec_CAF1 55503 119 - 1 AGCCC-GGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACCCAUCACACCGC ....(-((.(((((((((((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))........))))))...)))..))). ( -34.20) >DroSim_CAF1 60640 119 - 1 AGCCC-AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGC .....-.(.((.((((((((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))........)))))).)).)....... ( -33.00) >DroEre_CAF1 40811 119 - 1 GGCCA-AGCAGAGUGAUUAAUUAAUCAGAAUUUUCUGAUUGAUUUCGUCAAGGUUUUGGCUAAAAAAUGGCACCGCUACAACUGAACCUACACCACCAUCAAAAUCACGCAUCACACCGC ((...-......(((((((((((((((((....)))))))))))......((((((..((((.....))))............)))))).............))))))........)).. ( -23.67) >DroYak_CAF1 53620 120 - 1 AGCCAAAACCGAGAGCUUAAUUAAUCAGAAUUUCCCGAUUGAUUUCGUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGC .((.........((.(..((((((((.(......).))))))))..)))..((((((((.(((....((((...))))....))).))))))))..............)).......... ( -24.30) >consensus AGCCC_AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACAACUUAACCUACACCACCAUCAAAAUCACGCAUCACACCGC ..........(.((((((((((((((((......)))))))))).......((((((((.(((....((((...))))....))).))))))))........)))))).).......... (-26.36 = -27.56 + 1.20)



| Location | 6,796,980 – 6,797,088 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.24 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6796980 108 - 27905053 -----AAUGUCCUACUUA------UUAAACCCAAAAUGUAAGCCC-AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACA -----.........((((------(............)))))...-(((((((.((..((((((((((......)))))))))))))))).(((((.............))))))))... ( -23.52) >DroSec_CAF1 55543 119 - 1 UUAUUAAACUCCUACUUCAUACUCCUAAACCAAAUACUUAAGCCC-GGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACA .......................((((.(((.........(((..-.)))(((.((..((((((((((......)))))))))))))))..))).))))........((((...)))).. ( -23.00) >DroSim_CAF1 60680 113 - 1 UUAUUAAACUCCUACUUA------UUAAACCAAAGCCUUAAGCCC-AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACA ..................------.........(((((...(((.-...((((.((..((((((((((......)))))))))))))))).)))..)))))......((((...)))).. ( -26.10) >DroEre_CAF1 40851 108 - 1 -----AAUGUCAGACCUA------UUAAGCCCAACAGUUUGGCCA-AGCAGAGUGAUUAAUUAAUCAGAAUUUUCUGAUUGAUUUCGUCAAGGUUUUGGCUAAAAAAUGGCACCGCUACA -----.............------....(((......((((((((-(((....((((.(((((((((((....)))))))))))..))))..).))))))))))....)))......... ( -30.60) >DroYak_CAF1 53660 109 - 1 -----AAUGUCCAACUUA------UUAAGCUUGACAUUAAAGCCAAAACCGAGAGCUUAAUUAAUCAGAAUUUCCCGAUUGAUUUCGUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACA -----((((((...((..------...))...))))))..((((...(((..((.(..((((((((.(......).))))))))..)))..)))...))))......((((...)))).. ( -21.70) >consensus _____AAUGUCCUACUUA______UUAAACCAAACACUUAAGCCC_AGCUGAGUGAUUAAUUAAUCAGAAUUUCCUGAUUGAUUUCCUCAAGGUGUAGGCUAAAAAAUGGCACCGCUACA ........................................((((...(((...(((..((((((((((......))))))))))...))).)))...))))......((((...)))).. (-19.12 = -19.48 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:56 2006