
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,781,606 – 6,781,699 |
| Length | 93 |
| Max. P | 0.853946 |

| Location | 6,781,606 – 6,781,699 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6781606 93 + 27905053 GUUUUGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCAUC------GCCGACCGAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ((((..((((........))))))))...(((((((((...------((((........))))........(((.........)))...))))))))). ( -24.80) >DroSec_CAF1 45723 93 + 1 GUUUUGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCCUC------GCCGACCGAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ((((..((((........))))))))...(((((((((...------((((........))))........(((.........)))...))))))))). ( -23.90) >DroSim_CAF1 49757 93 + 1 GUUUUGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCCUC------GCCGACCGAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ((((..((((........))))))))...(((((((((...------((((........))))........(((.........)))...))))))))). ( -23.90) >DroEre_CAF1 30983 93 + 1 GUUUUGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCCUC------ACCGACCAAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ((((..((((........))))))))...(((((((((...------...(.(((....))))........(((.........)))...))))))))). ( -21.30) >DroYak_CAF1 43387 99 + 1 GUUUCGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCAUUUUGUUAGGCGACCGAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ((((..((((........))))))))...(((((((((..(((((((..(....)....))))))).....(((.........)))...))))))))). ( -23.60) >DroAna_CAF1 2838 88 + 1 GC----GUGUAAACAUGAGCACAAACAUUUUGUCAGCCCAU------GUCGGCU-AAAAUAGCAAAUAAAAUCAACAAAUAAAUGAAAAGCCUGACAAU ..----((((........)))).......(((((((....(------((..(((-(...))))..)))...(((.........))).....))))))). ( -13.90) >consensus GUUUUGGUGUAAACAUGGGCACGAACAUUUUGUCAGGCCUC______GCCGACCGAAAAUGGCAAAUAAAAUCAGCAAAUAAAUGAAAAGCCUGACAAU ......((((........)))).......(((((((((.........((((........))))........(((.........)))...))))))))). (-18.21 = -18.77 + 0.56)

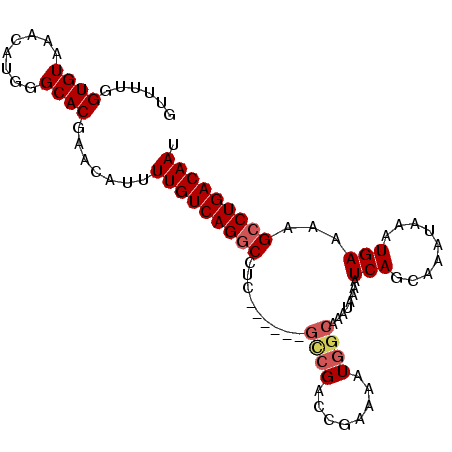

| Location | 6,781,606 – 6,781,699 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -21.43 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.56 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6781606 93 - 27905053 AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUCGGUCGGC------GAUGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCAAAAC .(((((((((...........(((((((((...............)))))))------)).)))))))))..(((.(((....)))...)))....... ( -22.16) >DroSec_CAF1 45723 93 - 1 AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUCGGUCGGC------GAGGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCAAAAC .((((((((((..........(((((((((...............)))))))------))))))))))))..(((.(((....)))...)))....... ( -23.36) >DroSim_CAF1 49757 93 - 1 AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUCGGUCGGC------GAGGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCAAAAC .((((((((((..........(((((((((...............)))))))------))))))))))))..(((.(((....)))...)))....... ( -23.36) >DroEre_CAF1 30983 93 - 1 AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUUGGUCGGU------GAGGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCAAAAC .((((((((((..(((.........)))..((((..((((....))))..))------))))))))))))..(((.(((....)))...)))....... ( -23.10) >DroYak_CAF1 43387 99 - 1 AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUCGGUCGCCUAACAAAAUGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCGAAAC .(((((((((.............(((((...............))))).............)))))))))....((((((..........)).)))).. ( -19.43) >DroAna_CAF1 2838 88 - 1 AUUGUCAGGCUUUUCAUUUAUUUGUUGAUUUUAUUUGCUAUUUU-AGCCGAC------AUGGGCUGACAAAAUGUUUGUGCUCAUGUUUACAC----GC .......((((..(((.........)))................-))))(((------((((((.(((.....)))...))))))))).....----.. ( -17.17) >consensus AUUGUCAGGCUUUUCAUUUAUUUGCUGAUUUUAUUUGCCAUUUUCGGUCGGC______GAGGCCUGACAAAAUGUUCGUGCCCAUGUUUACACCAAAAC .((((((((((((..........(((((...............)))))..........))))))))))))..(((.(((....)))...)))....... (-18.61 = -18.56 + -0.05)

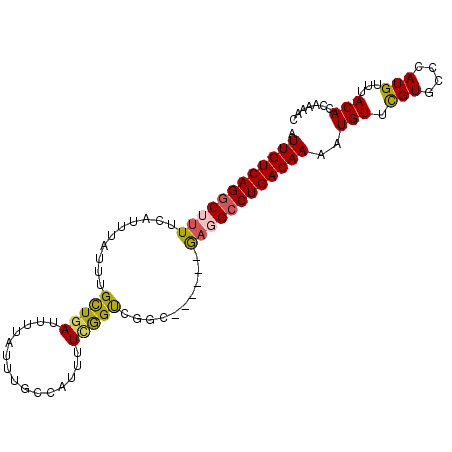
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:43 2006