
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,779,018 – 6,779,175 |
| Length | 157 |
| Max. P | 0.999671 |

| Location | 6,779,018 – 6,779,135 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.22 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -20.84 |
| Energy contribution | -20.76 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
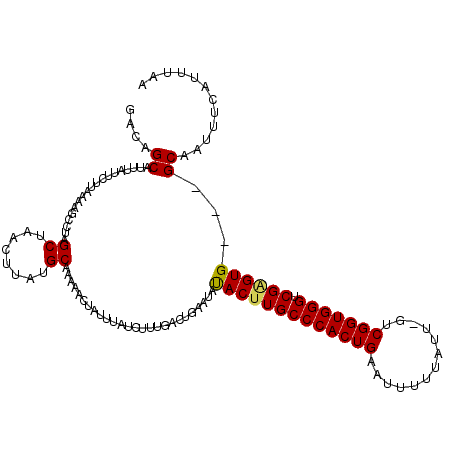
>3R_DroMel_CAF1 6779018 117 + 27905053 GGCAGCAUUUAUUCUCAAAAGCCUAGCUAACUUAUGCAAAAACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUCGGUCGGUGGGUCGAGUG---GCAAUUUCAUUUAA (((.................)))..((........))...............((((.((((..((((((((((((((...........)))))))).))))))---.....)))).)))) ( -25.43) >DroSec_CAF1 43171 116 + 1 GACAGCAUUUAUUUUUAAAAGCCUAGCUAACUUAUGCAAAAACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUU-GUCGGUGGGUCGAGUG---GCAAUUUCAUUUAA ....((.((((....)))).))...((........))...............((((.((((..((((((((((((((.........-.)))))))).))))))---.....)))).)))) ( -25.10) >DroSim_CAF1 47190 116 + 1 GACAGCAUUUAUUCUUAAAAGCCUUGCUAACUUAUGCAAAAACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUC-GUCGGUGGGUCGAGUG---GCAAUUUCAUUUAA ....((.((((....)))).)).((((........)))).............((((.((((..((((((((((((((.........-.)))))))).))))))---.....)))).)))) ( -26.60) >DroEre_CAF1 28235 115 + 1 AACCGCAUUUAUUCGUAAAAGCCGAGCUAACUUAUGCAAAAACUAUCUGUUUUUGACUGAAUAUAC-UGCCCACUGAAUUUUUAUU-GCCGGUGGGCCGAGUG---GCAAUUUCAUUUAA ..((((...(((((.....(((...)))........(((((((.....)))))))...)))))..(-.((((((((..........-..)))))))).).)))---)............. ( -28.20) >DroYak_CAF1 40548 119 + 1 GACAGCAGUUAUUCCUAAAAGCUACGCUAACUUAUGCAAAAACUAUUUAUCUUUAACUGAGUACACUUGCCCACUGAAUUUUUAUU-GCCGGUGGGCCGGGUGGCGGCAAUUUCAUUUAA .....((((((....((((((....((........)).....)).))))....)))))).((.(((((((((((((..........-..))))))).))))))...))............ ( -26.40) >consensus GACAGCAUUUAUUCUUAAAAGCCUAGCUAACUUAUGCAAAAACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUU_GUCGGUGGGUCGAGUG___GCAAUUUCAUUUAA ....((...................((........))..........................(((((((((((((.............))))))).))))))...))............ (-20.84 = -20.76 + -0.08)


| Location | 6,779,058 – 6,779,175 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -27.96 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
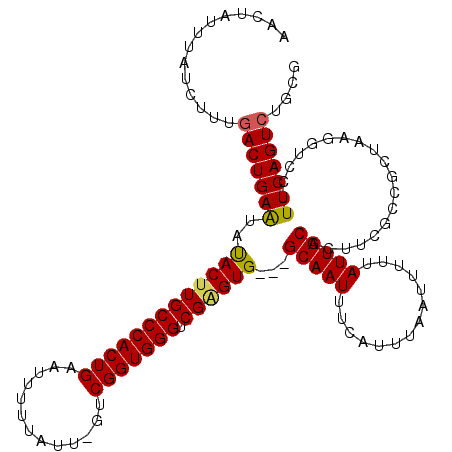
>3R_DroMel_CAF1 6779058 117 + 27905053 AACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUCGGUCGGUGGGUCGAGUG---GCAAUUUCAUUUAAUUUUUAUUGCACUUUGCCGCUAAGGUCCUUUCAGUCUGCG ..............(((((((...(((((((((((((...........)))))))))..((((---((((...((............))....)))))))).))))...))))))).... ( -34.30) >DroSec_CAF1 43211 115 + 1 AACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUU-GUCGGUGGGUCGAGUG---GCAAUUUCAUUUAAUUUUUAUUGCACUUCGCCGCUAAGG-CCUUUCAGUCUGCG ..............(((((((..((((((((((((((.........-.)))))))).))))))---(((((..............))))).....(((.....))-)..))))))).... ( -34.94) >DroSim_CAF1 47230 116 + 1 AACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUC-GUCGGUGGGUCGAGUG---GCAAUUUCAUUUAAUUUUUAUUGCACUUCGCCGCUAAGGUCCUUUCAGUCUGCG ..............(((((((..((((((((((((((.........-.)))))))).))))))---(((((..............))))).....(((.....)))...))))))).... ( -32.84) >DroEre_CAF1 28275 115 + 1 AACUAUCUGUUUUUGACUGAAUAUAC-UGCCCACUGAAUUUUUAUU-GCCGGUGGGCCGAGUG---GCAAUUUCAUUUAAUUUUUAUUGCACUUCUGCGCUAAGGUCCUUUCAGUCUGCC ..............(((((((...((-(((((((((..........-..)))))))).(((((---.((((..............))))))))).........)))...))))))).... ( -29.44) >DroYak_CAF1 40588 119 + 1 AACUAUUUAUCUUUAACUGAGUACACUUGCCCACUGAAUUUUUAUU-GCCGGUGGGCCGGGUGGCGGCAAUUUCAUUUAAUUUUUAUUGCACUUCUGCGCCAAGGUCCUUUCAGUCUGCG ...............((((((..(((((((((((((..........-..))))))).))))))..(((((((......))))......(((....))))))........))))))..... ( -30.00) >consensus AACUAUUUAUCUUUGACUGAAUAUACUUGCCCACUGAAUUUUUAUU_GUCGGUGGGUCGAGUG___GCAAUUUCAUUUAAUUUUUAUUGCACUUCGCCGCUAAGGUCCUUUCAGUCUGCG ..............(((((((..(((((((((((((.............))))))).))))))...(((((..............)))))...................))))))).... (-27.96 = -27.92 + -0.04)

| Location | 6,779,058 – 6,779,175 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.01 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6779058 117 - 27905053 CGCAGACUGAAAGGACCUUAGCGGCAAAGUGCAAUAAAAAUUAAAUGAAAUUGC---CACUCGACCCACCGACCGAUAAAAAUUCAGUGGGCAAGUAUAUUCAGUCAAAGAUAAAUAGUU ....(((((((...........(((((....((............))...))))---)(((.(.(((((.((...........)).)))))).)))...))))))).............. ( -24.90) >DroSec_CAF1 43211 115 - 1 CGCAGACUGAAAGG-CCUUAGCGGCGAAGUGCAAUAAAAAUUAAAUGAAAUUGC---CACUCGACCCACCGAC-AAUAAAAAUUCAGUGGGCAAGUAUAUUCAGUCAAAGAUAAAUAGUU ....(((((((..(-((.....)))((.(((((((..............)))).---)))))(.(((((.((.-.........)).)))))).......))))))).............. ( -26.34) >DroSim_CAF1 47230 116 - 1 CGCAGACUGAAAGGACCUUAGCGGCGAAGUGCAAUAAAAAUUAAAUGAAAUUGC---CACUCGACCCACCGAC-GAUAAAAAUUCAGUGGGCAAGUAUAUUCAGUCAAAGAUAAAUAGUU ....(((((((...........(((((....((............))...))))---)(((.(.(((((.((.-.........)).)))))).)))...))))))).............. ( -24.70) >DroEre_CAF1 28275 115 - 1 GGCAGACUGAAAGGACCUUAGCGCAGAAGUGCAAUAAAAAUUAAAUGAAAUUGC---CACUCGGCCCACCGGC-AAUAAAAAUUCAGUGGGCA-GUAUAUUCAGUCAAAAACAGAUAGUU ....(((((((.((......((((....))))......((((......)))).)---).....((((((.((.-.........)).)))))).-.....))))))).............. ( -26.30) >DroYak_CAF1 40588 119 - 1 CGCAGACUGAAAGGACCUUGGCGCAGAAGUGCAAUAAAAAUUAAAUGAAAUUGCCGCCACCCGGCCCACCGGC-AAUAAAAAUUCAGUGGGCAAGUGUACUCAGUUAAAGAUAAAUAGUU ....((((((....((((((((((....)))).............(((((((((((((....))).....)))-))).....)))).....)))).))..)))))).............. ( -26.30) >consensus CGCAGACUGAAAGGACCUUAGCGGCGAAGUGCAAUAAAAAUUAAAUGAAAUUGC___CACUCGACCCACCGAC_AAUAAAAAUUCAGUGGGCAAGUAUAUUCAGUCAAAGAUAAAUAGUU ....(((((((.........(((......)))..........................(((.(.(((((.((...........)).)))))).)))...))))))).............. (-19.54 = -19.74 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:34 2006