
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,777,947 – 6,778,167 |
| Length | 220 |
| Max. P | 0.998909 |

| Location | 6,777,947 – 6,778,065 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -34.79 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777947 118 + 27905053 UUUACGGCUGUCUCUCAGU--GUGCGCGCAAAGGAUGUGCGACCCAAGAAGUGUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUGGCACAGAAUCACGGCACAUGUGCAACAGCAAAA ......(((((..(...((--(((((((((.....)))))..........((((.(((...((((.....)))).........))).)))).........))))))..)..))))).... ( -33.00) >DroSec_CAF1 42130 117 + 1 UUUACGGCUGUCUCUCAGU--GUGCGCGCAAAGGAUGUGCGACCCAAGGAG-GUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUAGCACAGAAUCACGGCACAUGUGCAACAGCAAAA (((((..(((((((((..(--(.(((((((.....)))))).).))..)))-).)))))..))))).................(((.(((((.............))))).)))...... ( -34.02) >DroSim_CAF1 46110 117 + 1 UUUACGGCUGUCUCUCAGU--GUGCGCGCAAAGGAUGUGCGACCCAAGGAG-GUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUGGCACAGAAUCACGGCACAUGUGCAACAGCAAAA (((((..(((((((((..(--(.(((((((.....)))))).).))..)))-).)))))..))))).....((((........))))(((((.............))))).......... ( -35.22) >DroEre_CAF1 27245 117 + 1 UUUACGGCUGUCUCUCUGU--GUGCGCGCAAAGGAUGUGCGACCCAAGGAG-GUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUGGCACAAAAUCACGGCACAUGUGCAACAGCAAAA ......((..((((..((.--((.((((((.....)))))))).))..)))-)..))....((((.....))))......(((((((.(..........).)))))))((....)).... ( -35.70) >DroYak_CAF1 39539 119 + 1 UUUACGGCUGUCUCUCUGUGUGUGCGCGCAAAGGAUGUGCGACCCAAGGAG-GUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUGGCACAAAAUCACGGCACAUGUGCAACAGCAACA (((((..((((((((((.((.((.((((((.....)))))))).)).))))-).)))))..)))))..............(((((((.(..........).)))))))((....)).... ( -36.00) >consensus UUUACGGCUGUCUCUCAGU__GUGCGCGCAAAGGAUGUGCGACCCAAGGAG_GUGGCAGGAGUGAAAAAAUCACAGAAAAAUAUGUGGCACAGAAUCACGGCACAUGUGCAACAGCAAAA (((((..(((((.(((.....((.((((((.....)))))))).....)))...)))))..))))).....((((........))))(((((.............))))).......... (-28.76 = -29.16 + 0.40)



| Location | 6,777,947 – 6,778,065 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777947 118 - 27905053 UUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCACACUUCUUGGGUCGCACAUCCUUUGCGCGCAC--ACUGAGAGACAGCCGUAAA ....(((((..(.(((((((((..(...(((((.((..........((((.....)))).....(((.......))))).)))))....)..)).))))--).)).)..)))))...... ( -34.70) >DroSec_CAF1 42130 117 - 1 UUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCUACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCAC-CUCCUUGGGUCGCACAUCCUUUGCGCGCAC--ACUGAGAGACAGCCGUAAA ....(((((..(.(((((((((..(...(((((.((..........((((.....)))).....(((.-.....))))).)))))....)..)).))))--).)).)..)))))...... ( -35.20) >DroSim_CAF1 46110 117 - 1 UUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCAC-CUCCUUGGGUCGCACAUCCUUUGCGCGCAC--ACUGAGAGACAGCCGUAAA ....(((((..(.(((((((((..(...(((((.((..........((((.....)))).....(((.-.....))))).)))))....)..)).))))--).)).)..)))))...... ( -35.20) >DroEre_CAF1 27245 117 - 1 UUUUGCUGUUGCACAUGUGCCGUGAUUUUGUGCCACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCAC-CUCCUUGGGUCGCACAUCCUUUGCGCGCAC--ACAGAGAGACAGCCGUAAA ....((((((.....(((((((..(...(((((.((..........((((.....)))).....(((.-.....))))).)))))....)..)).))))--)......))))))...... ( -34.50) >DroYak_CAF1 39539 119 - 1 UGUUGCUGUUGCACAUGUGCCGUGAUUUUGUGCCACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCAC-CUCCUUGGGUCGCACAUCCUUUGCGCGCACACACAGAGAGACAGCCGUAAA ....((((((.....(((((((..(...(((((.((..........((((.....)))).....(((.-.....))))).)))))....)..)).)))))........))))))...... ( -33.32) >consensus UUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAUUUUUCUGUGAUUUUUUCACUCCUGCCAC_CUCCUUGGGUCGCACAUCCUUUGCGCGCAC__ACUGAGAGACAGCCGUAAA ....((((((.((...((((((..(...(((((.((..........((((.....)))).....(((.......))))).)))))....)..)).))))....))...))))))...... (-31.06 = -31.46 + 0.40)



| Location | 6,778,025 – 6,778,127 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6778025 102 + 27905053 AUAUGUGGCACAGAAUCACGGCACAUGUGCAACAGCAAAAGCA------------ACAGCAACAUUGGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUG (((((((.(..........).)))))))((....(((((.((.------------..........(((((((.......)))))))..((....)).)).)))))..))..... ( -26.40) >DroSim_CAF1 46187 102 + 1 AUAUGUGGCACAGAAUCACGGCACAUGUGCAACAGCAAAAGCA------------ACAGCAACAUUGGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUG (((((((.(..........).)))))))((....(((((.((.------------..........(((((((.......)))))))..((....)).)).)))))..))..... ( -26.40) >DroEre_CAF1 27322 114 + 1 AUAUGUGGCACAAAAUCACGGCACAUGUGCAACAGCAAAAGCAACAGCAAAGGCAACAGCAACAGUAGAAGCAACAUAAGCUUCCAGAGCAUCCGCAGCAUUUGUGAGCUAAUG .(((((.((...........(((....)))....((....((....))...(....).))..........)).)))))((((..((((((.......)).))))..)))).... ( -25.30) >DroYak_CAF1 39618 114 + 1 AUAUGUGGCACAAAAUCACGGCACAUGUGCAACAGCAACAGCAAAAGGAAAAGCAACAGCAACAGUAGAAGCAACAUAAGCUUCCAGAGCAUCCGCAGCAUUUGUGAGCUAAUG (((((((.(..........).)))))))((....((....))..........((....))..........))..(((.((((..((((((.......)).))))..)))).))) ( -23.80) >consensus AUAUGUGGCACAAAAUCACGGCACAUGUGCAACAGCAAAAGCA____________ACAGCAACAGUAGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUG (((((((.(..........).)))))))((....(((((.((.......................(((((((.......)))))))..((....)).)).)))))..))..... (-22.88 = -23.62 + 0.75)


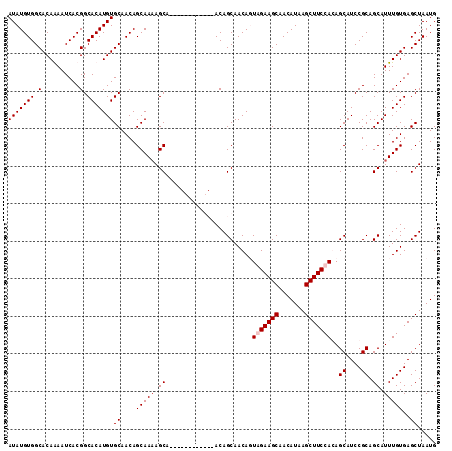
| Location | 6,778,025 – 6,778,127 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -25.15 |
| Energy contribution | -24.89 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6778025 102 - 27905053 CAUUAGCUCACAAAUGCUGCGGAUGCUGUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU------------UGCUUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAU .....((.((((((.((.((((..((..(((((((.......)))))))..))..))))------------.)).)))).))..))..(((((.((........)).))))).. ( -28.80) >DroSim_CAF1 46187 102 - 1 CAUUAGCUCACAAAUGCUGCGGAUGCUGUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU------------UGCUUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAU .....((.((((((.((.((((..((..(((((((.......)))))))..))..))))------------.)).)))).))..))..(((((.((........)).))))).. ( -28.80) >DroEre_CAF1 27322 114 - 1 CAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCUACUGUUGCUGUUGCCUUUGCUGUUGCUUUUGCUGUUGCACAUGUGCCGUGAUUUUGUGCCACAUAU ((..(((.((((((.((.((((..((..(((((((.......)))))))..))..)))).)).)))).))..)))..)).(((.(((((.............))))).)))... ( -33.02) >DroYak_CAF1 39618 114 - 1 CAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCUACUGUUGCUGUUGCUUUUCCUUUUGCUGUUGCUGUUGCACAUGUGCCGUGAUUUUGUGCCACAUAU ((.((((........((.((((..((..(((((((.......)))))))..))..)))).))..........)))).)).(((.(((((.............))))).)))... ( -28.29) >consensus CAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU____________UGCUUUUGCUGUUGCACAUGUGCCGUGAUUCUGUGCCACAUAU ((..(((...........((((..((..(((((((.......)))))))..))..)))).............)))..)).(((.(((((.............))))).)))... (-25.15 = -24.89 + -0.25)



| Location | 6,778,065 – 6,778,167 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6778065 102 + 27905053 GCA------------ACAGCAACAUUGGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUGACAUUAUAAACAUGAAAACUCCAUUAAUAUUUUUUUGCUG ...------------.((((((...(((((((.......))))))).(((...(((((...))))).)))......................................)))))) ( -20.40) >DroSim_CAF1 46227 102 + 1 GCA------------ACAGCAACAUUGGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUGACAUUAUAAACAUGAAAACUCCAUUAAUAUUUUUUUGCUG ...------------.((((((...(((((((.......))))))).(((...(((((...))))).)))......................................)))))) ( -20.40) >DroEre_CAF1 27362 114 + 1 GCAACAGCAAAGGCAACAGCAACAGUAGAAGCAACAUAAGCUUCCAGAGCAUCCGCAGCAUUUGUGAGCUAAUGACAUUAUAAACAUGAAAACUCCAUUAAUAUUUUUUUGCUG ....((((((((((..(.((....)).)..))..(((.((((..((((((.......)).))))..)))).)))................................)))))))) ( -23.30) >DroYak_CAF1 39658 114 + 1 GCAAAAGGAAAAGCAACAGCAACAGUAGAAGCAACAUAAGCUUCCAGAGCAUCCGCAGCAUUUGUGAGCUAAUGACAUUAUAAACAUGAAAACUCCAUUAAUAUUUUUUUGCUG (((((((((...((..(.((....)).)..))..(((.((((..((((((.......)).))))..)))).))).............................))))))))).. ( -22.80) >consensus GCA____________ACAGCAACAGUAGAAGCAACAUAAGCUUCCACAGCAUCCGCAGCAUUUGUGAGCUAAUGACAUUAUAAACAUGAAAACUCCAUUAAUAUUUUUUUGCUG ................((((((...(((((((.......))))))).(((...(((((...))))).)))......................................)))))) (-17.40 = -17.90 + 0.50)

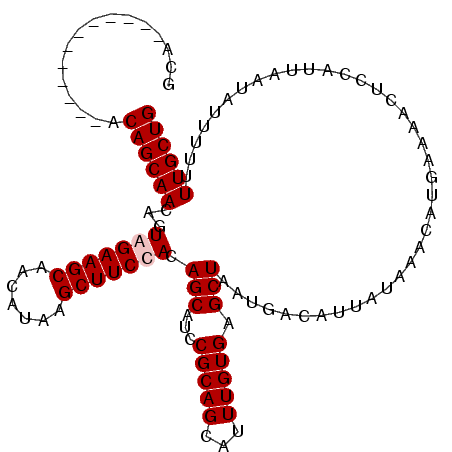

| Location | 6,778,065 – 6,778,167 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6778065 102 - 27905053 CAGCAAAAAAAUAUUAAUGGAGUUUUCAUGUUUAUAAUGUCAUUAGCUCACAAAUGCUGCGGAUGCUGUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU------------UGC ((((((....(((...(((((...)))))...)))...(((..((((........))))..)))....(((((((.......)))))))...)))))).------------... ( -24.00) >DroSim_CAF1 46227 102 - 1 CAGCAAAAAAAUAUUAAUGGAGUUUUCAUGUUUAUAAUGUCAUUAGCUCACAAAUGCUGCGGAUGCUGUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU------------UGC ((((((....(((...(((((...)))))...)))...(((..((((........))))..)))....(((((((.......)))))))...)))))).------------... ( -24.00) >DroEre_CAF1 27362 114 - 1 CAGCAAAAAAAUAUUAAUGGAGUUUUCAUGUUUAUAAUGUCAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCUACUGUUGCUGUUGCCUUUGCUGUUGC (((((((..........(((((((..(((.......))).....))))).))...((.((((..((..(((((((.......)))))))..))..)))).)).))))))).... ( -30.40) >DroYak_CAF1 39658 114 - 1 CAGCAAAAAAAUAUUAAUGGAGUUUUCAUGUUUAUAAUGUCAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCUACUGUUGCUGUUGCUUUUCCUUUUGC ..((((((.........(((((((..(((.......))).....))))).))...((.((((..((..(((((((.......)))))))..))..)))).))......)))))) ( -27.10) >consensus CAGCAAAAAAAUAUUAAUGGAGUUUUCAUGUUUAUAAUGUCAUUAGCUCACAAAUGCUGCGGAUGCUCUGGAAGCUUAUGUUGCUUCCAAUGUUGCUGU____________UGC ((((((....(((...(((((...)))))...)))...(((..((((........))))..)))....(((((((.......)))))))...))))))................ (-22.90 = -22.65 + -0.25)


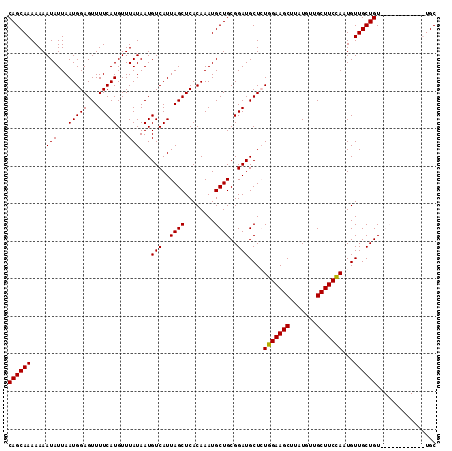
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:31 2006