
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,777,618 – 6,777,918 |
| Length | 300 |
| Max. P | 0.998938 |

| Location | 6,777,618 – 6,777,720 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777618 102 - 27905053 UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAGCACAUGAGCAGCAGUAACAACAA------AAACUUGUG-AAUCCGAAAAGGGGGCAGCUUGUAAAGCA-----------A ...((((((.....((((((...((((..((((((((((.....)))))....((....)).)------))))..)))-)......))))))....))))))......-----------. ( -23.70) >DroSec_CAF1 41792 108 - 1 UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGUUUAGCACAUGAGCAGCAACAACAACUACUGGGGUAGCUUGUG-AAUGCGAAAAGGGGGCGGCUUGUAAAGCA-----------A ...((((((.....((((((..(((((.((((......)))).)))))..(((...((((((((....)))).)))).-..)))..))))))....))))))......-----------. ( -33.60) >DroSim_CAF1 45772 108 - 1 UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAGCACAUGAGCAGCAACAACAACUACUGGGGUAGCUUGUG-AAUGCGAAAAGGGGGCGGCUUGUAAAGCA-----------A ...((((((.....((((((..(((((.((((......)))).)))))..(((...((((((((....)))).)))).-..)))..))))))....))))))......-----------. ( -33.60) >DroEre_CAF1 26886 111 - 1 UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAGCACAUGAGCAGCUACAACAA---------CAGCUACAGCAACAACCACUAGGGGCGGCUUGCAAAACAGGGGGCGUGUGA ...((((((.....((((....(((((.((((......)))).))))).((((.......---------.))))...............))))...))))))...(((.(...).))).. ( -25.40) >DroYak_CAF1 39184 113 - 1 UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUAGCUUAGCACAUGAGCAGCAACAACUG------GGGAAGCUGGUG-AAUGCGCAAAGGGGGCGGCUUGUAAAACAGGGGGCGUGCGA ...(((........((((((..(((((.((((......)))).)))))..(((...((..------(.....)..)).-..)))..)))))).((..((((.....))))..)).))).. ( -31.70) >consensus UUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAGCACAUGAGCAGCAACAACAAC_A___GGGAAGCUUGUG_AAUGCGAAAAGGGGGCGGCUUGUAAAGCA___________A ...((((((.....((((((..(((((.((((......)))).))))).(((...................))).............))))))...)))))).................. (-20.87 = -21.15 + 0.28)



| Location | 6,777,680 – 6,777,776 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -24.85 |
| Energy contribution | -24.89 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777680 96 - 27905053 GAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAG (((((((...((.(((((.((..(((.((((------------------------(...))))).)))..))((((.....))))......))))))))))))))............... ( -30.50) >DroSec_CAF1 41860 96 - 1 GAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGUUUAG (((((((...((.(((((.((..(((.((((------------------------(...))))).)))..))((((.....))))......))))))))))))))............... ( -30.50) >DroSim_CAF1 45840 96 - 1 GAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAG (((((((...((.(((((.((..(((.((((------------------------(...))))).)))..))((((.....))))......))))))))))))))............... ( -30.50) >DroEre_CAF1 26957 112 - 1 GAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------CUGGGUUGCUGGGUUGCUUGGCGACGGGGUGGCGCUACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAG (((((((...((.(((((.((..(((.((((--------(..(((.((...)).)))..))))).)))..))((((.....))))......))))))))))))))............... ( -39.40) >DroYak_CAF1 39257 119 - 1 GAGUUGGUUUGGUGGUUUGGCUGCUUGGCUGCUUCACUGCUUGGUGGCUGGGUUGCUU-GCGACGGGGUGGCGCCACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUAGCUUAG (((((((...(((((((.(((..((.(((.........))).))..)))(((((((((-(((((..((((....)))).))).)))))))...))))...))).))))...))))))).. ( -43.20) >consensus GAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG________________________CUUGGCGACGGGGUGGCGCUACCAGUUAGCAAGUAAAAACCCUUUAACUCAUUUGUUUUGCUUAG ((((((((((((((((...((..(((.((((.............................)))).)))..)))))))).(....)......)))))....)))))............... (-24.85 = -24.89 + 0.04)



| Location | 6,777,720 – 6,777,816 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -40.41 |
| Consensus MFE | -33.94 |
| Energy contribution | -34.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777720 96 + 27905053 CUGGUAGCGCCACCCCGUCGCCAAG------------------------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGUUGGGCCCCUUGGC .(((.....))).......((((((------------------------...((.((((((....((((((.((((((.....))...)))).))))))...)))))).))...)))))) ( -36.80) >DroSec_CAF1 41900 96 + 1 CUGGUAGCGCCACCCCGUCGCCAAG------------------------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUUCUGGGCCCCUUGGC .(((..(((......)))..)))..------------------------...(((((..(((...((((((.((((((.....))...)))).)))))).(((...))))))..))))). ( -34.70) >DroSim_CAF1 45880 96 + 1 CUGGUAGCGCCACCCCGUCGCCAAG------------------------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGC .(((.....))).......((((((------------------------...((.((((((....((((((.((((((.....))...)))).))))))...)))))).))...)))))) ( -39.20) >DroEre_CAF1 26997 112 + 1 CUGGUAGCGCCACCCCGUCGCCAAGCAACCCAGCAACCCAG--------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGC .(((.....))).......((((((...((((((......(--------(......))(((....((((((.((((((.....))...)))).))))))...)))))))))...)))))) ( -42.60) >DroYak_CAF1 39297 119 + 1 CUGGUGGCGCCACCCCGUCGC-AAGCAACCCAGCCACCAAGCAGUGAAGCAGCCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGC .(((((((........(.(..-..))......)))))))....((.(((..(((.((((((....((((((.((((((.....))...)))).))))))...)))))).)))..))).)) ( -48.74) >consensus CUGGUAGCGCCACCCCGUCGCCAAG________________________CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGC .((((.(((......))).)))).............................(((.(((((....((((((.((((((.....))...)))).))))))...))))))))(((....))) (-33.94 = -34.18 + 0.24)



| Location | 6,777,720 – 6,777,816 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -52.92 |
| Consensus MFE | -42.62 |
| Energy contribution | -43.70 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777720 96 - 27905053 GCCAAGGGGCCCAACAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAG ((((((.((((...(((((..(((((((((((..(.....)...)))))))))))...)))))...)))).------------------------)))))).....((((....)))).. ( -46.70) >DroSec_CAF1 41900 96 - 1 GCCAAGGGGCCCAGAAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAG ((((((.((((.((.((((..(((((((((((..(.....)...)))))))))))...)))).)).)))).------------------------)))))).....((((....)))).. ( -46.50) >DroSim_CAF1 45880 96 - 1 GCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG------------------------CUUGGCGACGGGGUGGCGCUACCAG ((((((.((((.(((((((..(((((((((((..(.....)...)))))))))))...))))))).)))).------------------------)))))).....((((....)))).. ( -53.10) >DroEre_CAF1 26997 112 - 1 GCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------CUGGGUUGCUGGGUUGCUUGGCGACGGGGUGGCGCUACCAG ((((((.((((((((((((..(((((((((((..(.....)...)))))))))))...(((.((...)).)--------)).)))))))))))).)))))).....((((....)))).. ( -58.70) >DroYak_CAF1 39297 119 - 1 GCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGCUGCUUCACUGCUUGGUGGCUGGGUUGCUU-GCGACGGGGUGGCGCCACCAG ((.(((.((((.(((((((..(((((((((((..(.....)...)))))))))))...))))))).)))).)))....)).(((((((...((..(((-......)))..))))))))). ( -59.60) >consensus GCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG________________________CUUGGCGACGGGGUGGCGCUACCAG ((((((.((((.(((((((..(((((((((((..(.....)...)))))))))))...))))))).)))).........((((....))))....)))))).....((((....)))).. (-42.62 = -43.70 + 1.08)



| Location | 6,777,745 – 6,777,855 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -46.32 |
| Consensus MFE | -34.10 |
| Energy contribution | -35.34 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777745 110 + 27905053 ---------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGUUGGGCCCCUUGGCUUUAUGG-GGACCAUGGUGAAGCUGCUGGAGCUGCUAGAG ---------......(((((((...((((((.((((((.....))...)))).))))))((((((..(((.((((..........))-)).)))......))))))..).)))))).... ( -46.10) >DroSec_CAF1 41925 111 + 1 ---------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUUCUGGGCCCCUUGGCUUUAUGGGGGACCAUGGUGGAGCUGCUGAAGCCGCUGGAG ---------...(((.((.((....((((((.((((((.....))...)))).))))))((((((((..((((....))))((((((....)))))).))))))))....)).))))).. ( -49.00) >DroSim_CAF1 45905 111 + 1 ---------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGCUUUAUGGGGGAGCAUGGCGGAGCUGCUGGAGCCGCUGGAG ---------...(((.(((((....((((((.((((((.....))...)))).))))))...))))))))((((((((......)))))).))..(((((..(.....)..))))).... ( -46.30) >DroEre_CAF1 27037 111 + 1 G--------CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGCUUUAUGG-AGACCAUGGUGGAGCUGCUGGAGCCGCUGGAG .--------...(((((((((....((((((.((((((.....))...)))).))))))...)))))).(((((..((((((((((.-...)....)))))))))..)).)))..))).. ( -44.70) >DroYak_CAF1 39336 110 + 1 GCAGUGAAGCAGCCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGCUUUAUGG-AGACCAUGGUGGCGCUGGUG---------GAG .((((.......(((.(((((....((((((.((((((.....))...)))).))))))...))))))))(((((.((((((....)-)).))).)).)))))))...---------... ( -45.50) >consensus _________CAACCAAGCAGCCAAACCACCAAACCAACUCAUUGUGCAUGGUCUGGUGGGCAGCUGCUGGGCCCCUUGGCUUUAUGG_GGACCAUGGUGGAGCUGCUGGAGCCGCUGGAG ............(((.(((((....((((((.((((((.....))...)))).))))))...))))))))(((....))).((((((....))))))...(((.((....)).))).... (-34.10 = -35.34 + 1.24)

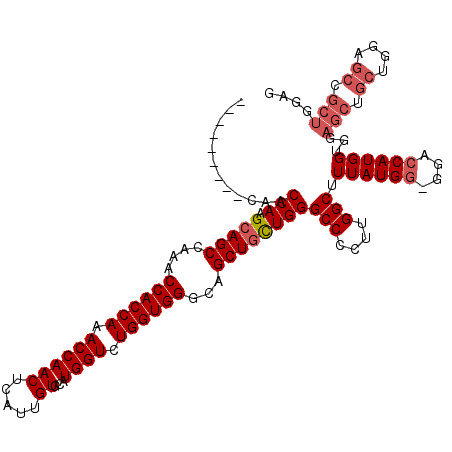
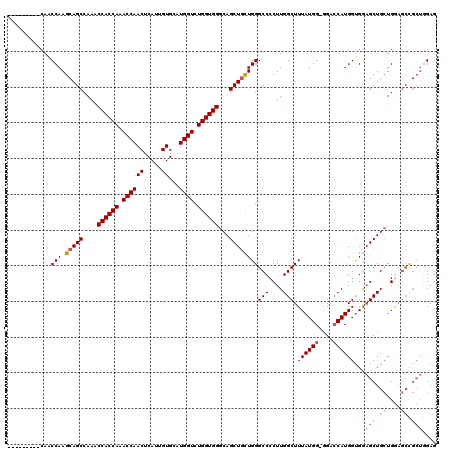
| Location | 6,777,745 – 6,777,855 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -47.56 |
| Consensus MFE | -38.62 |
| Energy contribution | -40.06 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777745 110 - 27905053 CUCUAGCAGCUCCAGCAGCUUCACCAUGGUCC-CCAUAAAGCCAAGGGGCCCAACAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------- .((.(((((((...((((((......(((.((-((..........)))).)))..))))))(((((((((((..(.....)...)))))))))))...))))))).))...--------- ( -47.70) >DroSec_CAF1 41925 111 - 1 CUCCAGCGGCUUCAGCAGCUCCACCAUGGUCCCCCAUAAAGCCAAGGGGCCCAGAAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------- ..(((((((((...((((((......(((.((((...........)))).)))..))))))(((((((((((..(.....)...)))))))))))...)))))).)))...--------- ( -49.20) >DroSim_CAF1 45905 111 - 1 CUCCAGCGGCUCCAGCAGCUCCGCCAUGCUCCCCCAUAAAGCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------- ..(((((((((...((((((..((...(((((.............)))))...))))))))(((((((((((..(.....)...)))))))))))...)))))).)))...--------- ( -47.12) >DroEre_CAF1 27037 111 - 1 CUCCAGCGGCUCCAGCAGCUCCACCAUGGUCU-CCAUAAAGCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG--------C ..(((((((((...((((((......(((.((-((..........)))).)))..))))))(((((((((((..(.....)...)))))))))))...)))))).)))...--------. ( -46.80) >DroYak_CAF1 39336 110 - 1 CUC---------CACCAGCGCCACCAUGGUCU-CCAUAAAGCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGCUGCUUCACUGC ...---------...(((((((.((.(((.((-......))))).)))))(.(((((((..(((((((((((..(.....)...)))))))))))...))))))).)))))......... ( -47.00) >consensus CUCCAGCGGCUCCAGCAGCUCCACCAUGGUCC_CCAUAAAGCCAAGGGGCCCAGCAGCUGCCCACCAGACCAUGCACAAUGAGUUGGUUUGGUGGUUUGGCUGCUUGGUUG_________ ....(((.((....)).)))...((.((((..........)))).))((((.(((((((..(((((((((((..(.....)...)))))))))))...))))))).)))).......... (-38.62 = -40.06 + 1.44)



| Location | 6,777,816 – 6,777,918 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -20.44 |
| Energy contribution | -22.68 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6777816 102 - 27905053 UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUCCAGCGGCUCUAGCAGCUCCAGCAGCUUCACCAUGGUCC-CCAUAAA ....(((((((..(((....)))))))))).(((.((((((((..((..((((.((..(((.((....)).)))..)).)))).))..)))))))-).))).. ( -33.80) >DroSec_CAF1 41996 103 - 1 UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUCCAGCGGCUCCAGCGGCUUCAGCAGCUCCACCAUGGUCCCCCAUAAA ......(((((..(((....))))))))(((((..((((((((..((..((((.((..(((.((....)).)))..)).)))).))..)))))))).))))). ( -35.10) >DroSim_CAF1 45976 103 - 1 UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUACGGCUUCUCCAGCGGCUCCAGCAGCUCCGCCAUGCUCCCCCAUAAA ....(((((((..(((....)))))))))).((((((((((.....((((.......)))).........))))))))))((........))........... ( -31.94) >DroEre_CAF1 27109 99 - 1 UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUCUGG---CUCCAGCGGCUCCAGCAGCUCCACCAUGGUCU-CCAUAAA ....(((((((..(((....)))))))))).((((((((((.....(((((.....))))---)......))))))))))........((((...-))))... ( -33.30) >DroYak_CAF1 39416 90 - 1 UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUCUGG---CUC---------CACCAGCGCCACCAUGGUCU-CCAUAAA ....(((((((..(((....)))))))))).(((.((((((((......(((....((((---...---------..)))).)))...)))))))-).))).. ( -25.60) >consensus UAUAAAUGUGCGAACACUAAUGUGCACAUUAUGUUGGGGCCAUUAUGCCGGCUUCUCCGGC__CUCCAGCGGCUCCAGCAGCUCCACCAUGGUCC_CCAUAAA ....(((((((..(((....)))))))))).((((((((((.....(((((.....))))).........))))))))))........((((....))))... (-20.44 = -22.68 + 2.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:26 2006