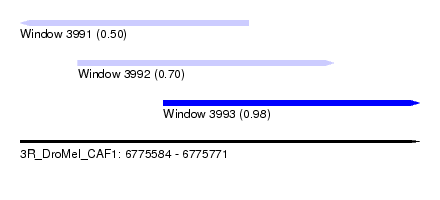
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,775,584 – 6,775,771 |
| Length | 187 |
| Max. P | 0.984561 |

| Location | 6,775,584 – 6,775,691 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6775584 107 - 27905053 UGCGGAAGACAUCUUCAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGAAAAUGGAUUGUGCUAAUUCAUGUCAGGAUCUGAAUCCCCCUUUUC-------------CCAUCCCUUAAC ...((.(((((.(((..(((((...)))))..))).))))).))....((..((((((((...)))))))).)).((((..(((........)))-------------..))))...... ( -23.60) >DroSec_CAF1 39764 107 - 1 UAAGGAAGACAUCUACAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGAAAAUGGAUUGUGCUAAUUCAUGUCAGAAUCUGAAACCCCAUUUUC-------------CCAUCCCUGAAC ..(((.(((((.((...(((((...)))))...)).))))).)))...((((((((......(..((((......))))..).....))))))))-------------............ ( -24.60) >DroSim_CAF1 43771 107 - 1 UAUGGAAGACAUCUACAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGAAAAUGGAUUGUGCUAAUUCAUGUCAGAAUCUGAAACCCCAUUUUC-------------CCAUCCCUGAAC .((((.(((((.((...(((((...)))))...)).))))).......((((((((......(..((((......))))..).....))))))))-------------))))........ ( -23.40) >DroEre_CAF1 24893 120 - 1 UAUGGAAGACAUCUUCAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGAAAAUGAAUUGUGCUAAUUCACGUCAGAAUCUUAAAUCUCCUUUUCGCCCUCCAAAAACCCAUUCCUUAAC ..(((((((((.(((..(((((...)))))..))).))))).....((((...(((...(((......))).)))...)))).................))))................. ( -21.40) >DroYak_CAF1 37111 120 - 1 UAUGGAAGACAGCUUCAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGCAAAUGGAUUGUGCUAAUUCAUGUCAGAAUCUGAAACUCCAUUUUCGCCCUUCUAAAACCCAUCCCUUAGC .((((.(((((((((..(((((...)))))..))))))))).......(((((((((.(...(..((((......))))..)..).)))))))..))...........))))........ ( -28.30) >consensus UAUGGAAGACAUCUUCAUUGAGCAGCUCAAGAAAGUUGUCUGCCUAAAGAAAAUGGAUUGUGCUAAUUCAUGUCAGAAUCUGAAACCCCAUUUUC_____________CCAUCCCUUAAC ...((.(((((.(((..(((((...)))))..))).))))).))....((((((((......(..((((......))))..).....))))))))......................... (-19.96 = -20.68 + 0.72)

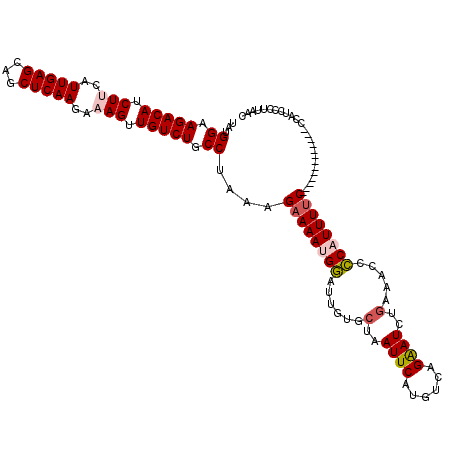
| Location | 6,775,611 – 6,775,731 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6775611 120 + 27905053 GAUCCUGACAUGAAUUAGCACAAUCCAUUUUCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCGCAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGAC ..............((((((.....(((((((.((..((((.(((.....)))..))))..)).))))))).(((((((....))).))))......................)))))). ( -24.60) >DroSec_CAF1 39791 120 + 1 GAUUCUGACAUGAAUUAGCACAAUCCAUUUUCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGUAGAUGUCUUCCUUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGAC (((((......))))).......(((((((.....(((.(((((.((...(((((...)))))...)).))))).)))...))))).))(((((.(((((......))))).)))))... ( -24.10) >DroSim_CAF1 43798 120 + 1 GAUUCUGACAUGAAUUAGCACAAUCCAUUUUCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGUAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGAC (((((......))))).......(((((((......((.(((((.((...(((((...)))))...)).))))).))....))))).))(((((.(((((......))))).)))))... ( -21.80) >DroEre_CAF1 24933 110 + 1 GAUUCUGACGUGAAUUAGCACAAUUCAUUUUCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUA----------AACUUGCUGAC ...(((.(((((((((.....)))))))........((.(((((.((((.(((((...))))).)))).))))).))......)).)))(((((.....----------...)))))... ( -26.10) >DroYak_CAF1 37151 119 + 1 GAUUCUGACAUGAAUUAGCACAAUCCAUUUGCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGAAGCUGUCUUCCAUAUAAGUGAGAAGUAACUUUA-AAUACAUUAAACGUGCUGCC (((((......)))))(((((......((..(((..((.(((((.((((.(((((...))))).)))).))))).))....)))..))..(((......-..))).......)))))... ( -29.60) >consensus GAUUCUGACAUGAAUUAGCACAAUCCAUUUUCUUUAGGCAGACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGAC (((((......)))))((((................((.(((((.((((.(((((...))))).)))).))))).))....((((........))))................))))... (-20.86 = -21.46 + 0.60)

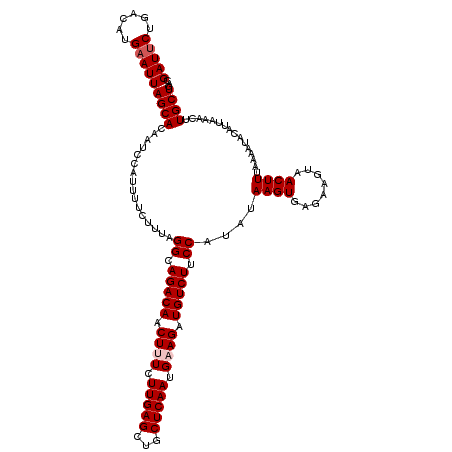
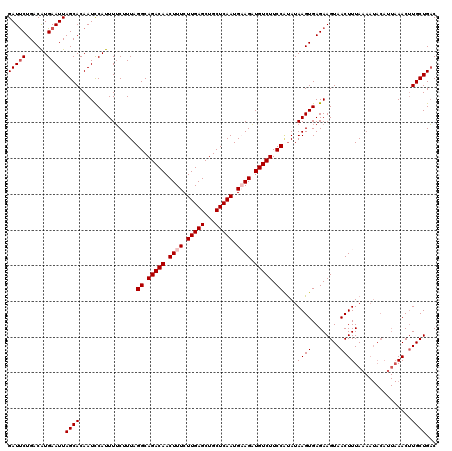
| Location | 6,775,651 – 6,775,771 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.41 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -20.87 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6775651 120 + 27905053 GACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCGCAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGACAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAUAAUAUU ((((.((((.(((((...))))).)))).))))..........((((((((((((((.............(((((((.........)))))))....))))).)).)).)))))...... ( -23.63) >DroSec_CAF1 39831 119 + 1 GACAACUUUCUUGAGCUGCUCAAUGUAGAUGUCUUCCUUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGACAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAUAAUAU- ((((.((...(((((...)))))...)).))))..........((((((((((((((.............(((((((.........)))))))....))))).)).)).))))).....- ( -20.43) >DroSim_CAF1 43838 120 + 1 GACAACUUUCUUGAGCUGCUCAAUGUAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGACAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAAAAUAUU ..........(((((((((.....))))(((.((((((((((.((....(((((.(((((......))))).))))).)).))))))).))).))).............)))))...... ( -24.40) >DroEre_CAF1 24973 110 + 1 GACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUA----------AACUUGCUGACAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAUAAUAUU ((((.((((.(((((...))))).)))).))))..........((((((((((((((..----------.(((((((.........)))))))....))))).)).)).)))))...... ( -27.00) >DroYak_CAF1 37191 119 + 1 GACAACUUUCUUGAGCUGCUCAAUGAAGCUGUCUUCCAUAUAAGUGAGAAGUAACUUUA-AAUACAUUAAACGUGCUGCCAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAUAAUAUU ((((.((((.(((((...))))).)))).))))..........(((((.((((..((((-(.....)))))..))))((((....))))....................)))))...... ( -24.50) >consensus GACAACUUUCUUGAGCUGCUCAAUGAAGAUGUCUUCCAUAUAAGUGAGAAGUAACUUUAAAAUACAUUAAACUUGCUGACAUAUAUGGCAAGUCAUUAAGUUCACAUUACUCAUAAUAUU ((((.((((.(((((...))))).)))).))))..........((((((((((((((.............(((((((.........)))))))....))))).)).)).)))))...... (-20.87 = -21.67 + 0.80)

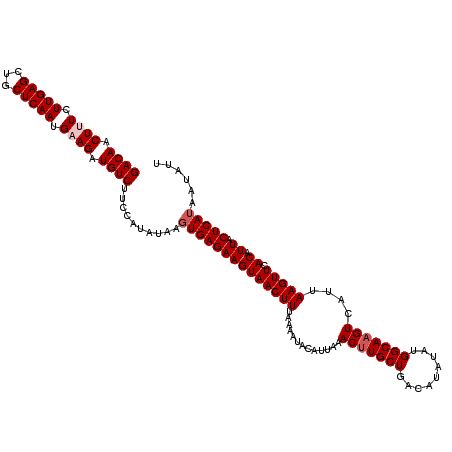
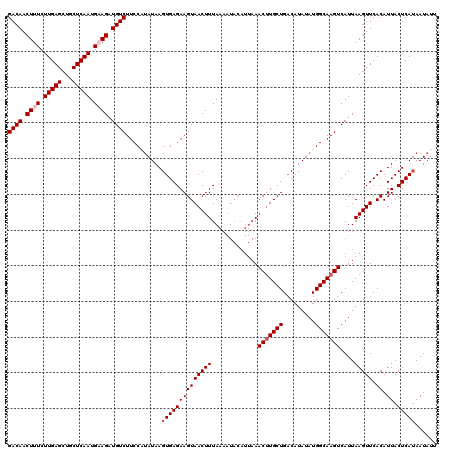
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:18 2006