
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,773,196 – 6,773,353 |
| Length | 157 |
| Max. P | 0.821431 |

| Location | 6,773,196 – 6,773,313 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -29.71 |
| Energy contribution | -31.03 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6773196 117 - 27905053 GCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCACACUUGUGGC---GGGCGGGAGGGGGAGGAGAGCAAAUAAUUUUUAAUUGUGCUGCACAAUGCUGUAAA ((((((......((((((((((((......))).)))))....((..(.(((....))).)---..)).))))..........(((.(((((.....)))))..)))....))))))... ( -38.10) >DroSec_CAF1 37442 117 - 1 GCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCACACUUGCGGA---GGGGCGGAGCGGGAGGAGAGCAAAUAAUUUUUAAUUGUGCUGCACAAUGCUGUAAA ((((((.....(((((...(((((......)))))((((....(((((.....)))))...---.))))))))).........(((.(((((.....)))))..)))....))))))... ( -43.80) >DroEre_CAF1 22476 111 - 1 GCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCA-AAGCAAGGCAAAGU-ACGGC---GGGGCGG---GGG-GGAGAGCAAAUAAUUUUUAAUUGUGCUGCACAAUGCUGUAAA ((((((.....(((((((((((((......))).))))).-..((...((...))-...))---)))))..---...-.....(((.(((((.....)))))..)))....))))))... ( -34.30) >DroYak_CAF1 34624 116 - 1 GCAGCAAUUAAGCUUCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCAAAGU-GUUGCAAGGGGGCGG---GGGAGGAGAGCAAAUAAUUUUUAAUUGUGCUGCACAAUGCUGUAAA ((((((.....(((((((((((((......))).)))))....((((.((...))-.))))...)))))..---.........(((.(((((.....)))))..)))....))))))... ( -35.10) >consensus GCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCAAACU_GCGGC___GGGGCGG___GGGAGGAGAGCAAAUAAUUUUUAAUUGUGCUGCACAAUGCUGUAAA ((((((.....(((((((((((((......))).)))))....(((((.....)))))......)))))..............(((.(((((.....)))))..)))....))))))... (-29.71 = -31.03 + 1.31)

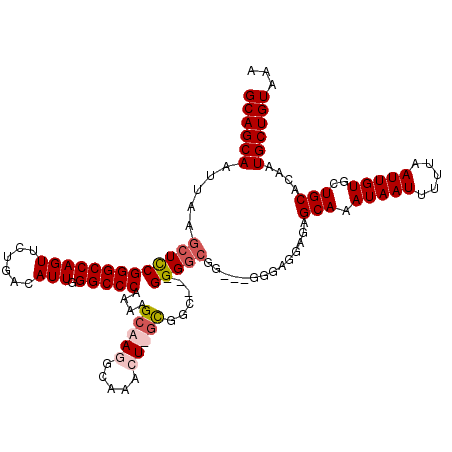
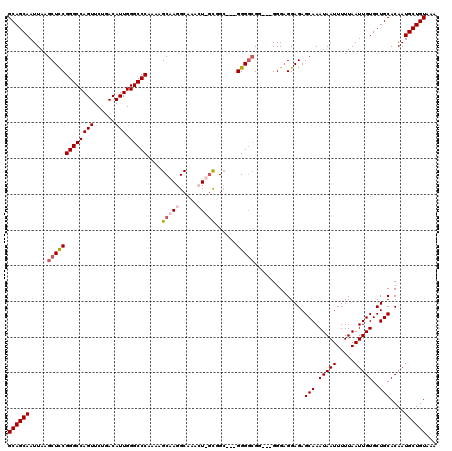
| Location | 6,773,236 – 6,773,353 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -32.45 |
| Energy contribution | -32.33 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6773236 117 + 27905053 CCUCCCCCUCCCGCCC---GCCACAAGUGUGCCUUGCUUUUGGGCCCAAUGUCAGAACUGGCCCGGAGCUUAAUUGCUGCAAAGUUGUUGUGGCUUAGCCGGUUGGGCGUGGCAGUAGGA ..(((..(((((((((---(((((((.(.(((...((..((((((((...((((....))))..)).))))))..)).))).).))))...(((...))))).)))))).)).))..))) ( -45.40) >DroSec_CAF1 37482 112 + 1 CCUCCCGCUCCGCCCC---UCCGCAAGUGUGCCUUGCUUUUGGGCCCAAUGUCAGAACUGGCCCGGAGCUUAAUUGCUGCAAAGUUGUUGUGGCUUAGCCGGUUGGGCGUGGCAG----- ......((..(((((.---.(((((((.....))))).((((((((....((....)).))))))))(((.....((..(((.....)))..))..))).))..)))))..))..----- ( -45.10) >DroEre_CAF1 22516 111 + 1 CC-CCC---CCGCCCC---GCCGU-ACUUUGCCUUGCUU-UGGGCCCAAUGUCAGAACUGGCCCGGAGCUUAAUUGCUGCAAAGUUGUUGUGGCUUAGCCGGUUGGGCGUGGCACUAGCA ..-..(---((((((.---((((.-.((..(((..((((-((((((....((....)).)))))))))).((((.(((....))).)))).)))..)).)))).))))).))........ ( -45.30) >DroYak_CAF1 34664 116 + 1 CCUCCC---CCGCCCCCUUGCAAC-ACUUUGCCUUGCUUUUGGGCCCAAUGUCAGAACUGGCCCGAAGCUUAAUUGCUGCAAAGUUGUUGUGGCUUAGCCGGUUGGGCGUGGCAGUAGCA .((.((---.(((((((..(((((-(((((((.......(((((((....((....)).)))))))(((......))))))))).))))))(((...)))))..))))).)).))..... ( -44.70) >consensus CCUCCC___CCGCCCC___GCCAC_ACUGUGCCUUGCUUUUGGGCCCAAUGUCAGAACUGGCCCGGAGCUUAAUUGCUGCAAAGUUGUUGUGGCUUAGCCGGUUGGGCGUGGCAGUAGCA ...................(((((......((((.......)))).....(((..(((((((...(((((((((.(((....))).)))).))))).))))))).))))))))....... (-32.45 = -32.33 + -0.13)

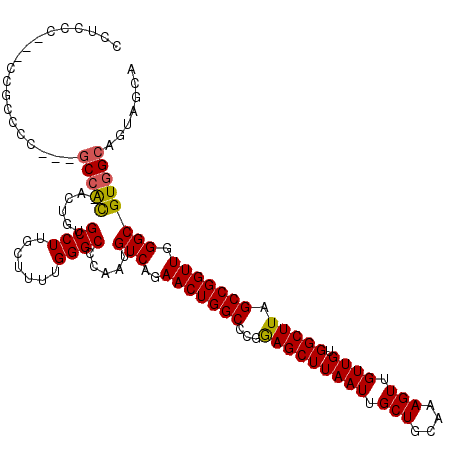
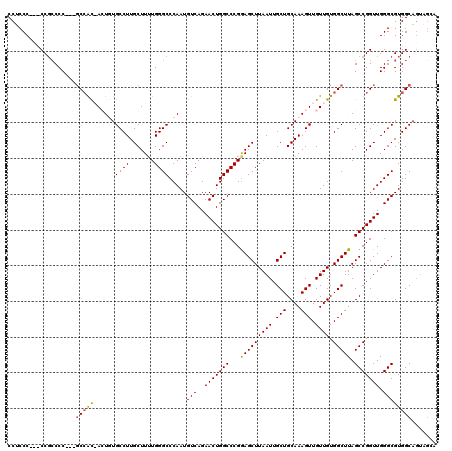
| Location | 6,773,236 – 6,773,353 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -45.23 |
| Consensus MFE | -33.38 |
| Energy contribution | -35.88 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6773236 117 - 27905053 UCCUACUGCCACGCCCAACCGGCUAAGCCACAACAACUUUGCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCACACUUGUGGC---GGGCGGGAGGGGGAGG ((((.(((((..(((.....)))...(((((((...(((((((((......)))..((((((((......))).)))))....)))))).....)))))))---.)))))....)))).. ( -47.20) >DroSec_CAF1 37482 112 - 1 -----CUGCCACGCCCAACCGGCUAAGCCACAACAACUUUGCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCACACUUGCGGA---GGGGCGGAGCGGGAGG -----((((..(((((....(((...))).......................((((((((((((......))).)))))....(((((.....))))))))---))))))..)))).... ( -45.60) >DroEre_CAF1 22516 111 - 1 UGCUAGUGCCACGCCCAACCGGCUAAGCCACAACAACUUUGCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCA-AAGCAAGGCAAAGU-ACGGC---GGGGCGG---GGG-GG ........((.(((((..(((((...)))......((((((((((......)))..((((((((......))).))))).-.......)))))))-..)).---.))))).---)).-.. ( -42.20) >DroYak_CAF1 34664 116 - 1 UGCUACUGCCACGCCCAACCGGCUAAGCCACAACAACUUUGCAGCAAUUAAGCUUCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCAAAGU-GUUGCAAGGGGGCGG---GGGAGG .....((.((.(((((..(((((...))).(((((.(((((((((......)))..((((((((......))).))))).........)))))))-))))...))))))).---)).)). ( -45.90) >consensus UGCUACUGCCACGCCCAACCGGCUAAGCCACAACAACUUUGCAGCAAUUAAGCUCCGGGCCAGUUCUGACAUUGGGCCCAAAAGCAAGGCAAACU_GCGGC___GGGGCGG___GGGAGG ........((...(((..(((.((..((((((((..(((((((((......)))..((((((((......))).)))))....))))))....))))))))....)).)))...))).)) (-33.38 = -35.88 + 2.50)

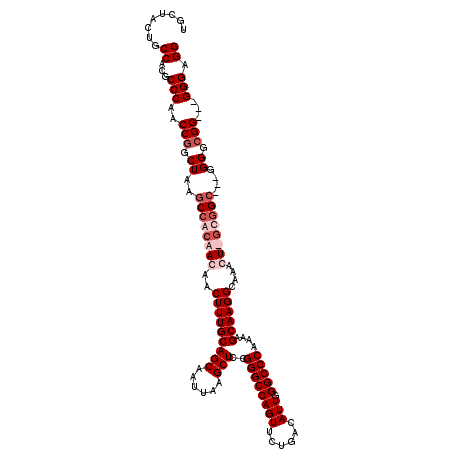
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:36:09 2006