| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
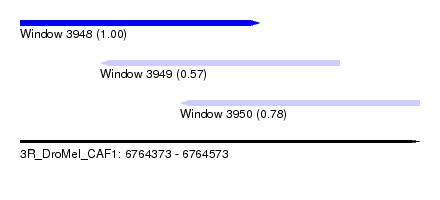
| Location | 6,764,373 – 6,764,573 |
| Length | 200 |
| Max. P | 0.996400 |

| Location | 6,764,373 – 6,764,493 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6764373 120 + 27905053 AUUUUAUCUCUCUUCAUCUCUUUAGUUUCGGCUUUCUGGGGUGAGAAACAACUUUUCAUUUUUGUCUCGGGCGCACAGUUUGACACCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU .........(((.(((((((...(((....)))....)))))))((.((((..........)))).))))).(((((.((((((((.....(((....))))))))))).)))))..... ( -30.00) >DroSec_CAF1 28503 120 + 1 AUUUUAUCUCUCUUCAUCUCUUUAGUUUCGGCUUUCUGGGGUGAGGAACAACUUUUCAUUUUUGUCUCGAGCGCACAGUUUGACACCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU .......(((((((((((((...(((....)))....))))))))))((((..........))))...))).(((((.((((((((.....(((....))))))))))).)))))..... ( -33.20) >DroSim_CAF1 33030 120 + 1 AUUUUAUCUCUUUUCAUCUCUUUAGUUUCGGCUUUCUGGGGUGAGGAACAACUUUUCAUUUUUGUCUCGGGCGCACAGUUUGACAUCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU .......(((...(((((((...(((....)))....)))))))(..((((..........))))..)))).(((((.((((((((.....(((....))))))))))).)))))..... ( -29.10) >DroEre_CAF1 13293 120 + 1 AUUUUAUCUCUCUUCGUCUCUUCAGUUCCGGCCUUUUGGGGUGAGGAACAACUUUUCAUUUUUGUCUCGGGUGCACAGUUUGACACCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU ........................(((((.((((....))))..))))).....................(((((((.((((((((.....(((....))))))))))).)))))))... ( -34.40) >DroYak_CAF1 25330 120 + 1 AUUUUAUCUCUCUUCAUCUCUUUAGUCCCGGCCUUUUGGGGUGAGGAACAACUUUUCAUUUUUGUCUCGAGCGCACAGUUUGACACCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU .......(((((((((((((....((....)).....))))))))))((((..........))))...))).(((((.((((((((.....(((....))))))))))).)))))..... ( -32.00) >consensus AUUUUAUCUCUCUUCAUCUCUUUAGUUUCGGCUUUCUGGGGUGAGGAACAACUUUUCAUUUUUGUCUCGGGCGCACAGUUUGACACCCGGUGUCAAAUGAUGUGUCAAAAUGUGCACAAU .......(((((((((((((...(((....)))....))))))))))((((..........))))...))).(((((.((((((((.....(((....))))))))))).)))))..... (-29.50 = -29.42 + -0.08)



| Location | 6,764,413 – 6,764,533 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -31.11 |
| Energy contribution | -30.75 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6764413 120 - 27905053 UGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCAAACUGUGCGCCCGAGACAAAAAUGAAAAGUUGUUUCUCAC ((((.....))))((.....)).((.(((.....)))......(((((((.((((((((.((..........)))))))))).)))))))))(((((((...(....).))))))).... ( -33.40) >DroSec_CAF1 28543 120 - 1 UGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGCAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCAAACUGUGCGCUCGAGACAAAAAUGAAAAGUUGUUCCUCAC ........(((((((((((.....(((((((.....)).....(((((((.((((((((.((..........)))))))))).))))))).)).))).......))))))))..)))... ( -33.70) >DroSim_CAF1 33070 120 - 1 UGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGAUGUCAAACUGUGCGCCCGAGACAAAAAUGAAAAGUUGUUCCUCAC ........(((((((((((.....((((((.............(((((((.(((((((.....((((....))))))))))).)))))))))).))).......))))))))..)))... ( -30.01) >DroEre_CAF1 13333 120 - 1 UGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCAAACUGUGCACCCGAGACAAAAAUGAAAAGUUGUUCCUCAC ........(((((((((((.....((((((.............(((((((.((((((((.((..........)))))))))).)))))))))).))).......))))))))..)))... ( -33.31) >DroYak_CAF1 25370 120 - 1 CGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCAAACUGUGCGCUCGAGACAAAAAUGAAAAGUUGUUCCUCAC ........(((((((((((.....(((((((..((((....))))(((((.((((((((.((..........)))))))))).))))))).)).))).......))))))))..)))... ( -32.10) >consensus UGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCAAACUGUGCGCCCGAGACAAAAAUGAAAAGUUGUUCCUCAC ........(((((((((((.....((((((.............(((((((.((((((((.((..........)))))))))).)))))))))).))).......))))))))..)))... (-31.11 = -30.75 + -0.36)



| Location | 6,764,453 – 6,764,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.23 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.28 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6764453 120 - 27905053 CAGCAGCCCACAGCUACCACCCACUGGCCACCUGCCCACCUGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCA .(((........)))..(((((....((((((((((.(((((.....))))).)......))))))..)))....(((....((((((.....)).))))((....)).))))))))... ( -33.70) >DroSec_CAF1 28583 112 - 1 CAGCAGCCCACAGCAACCACCCACU--------GCCCACCUGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGCAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCA ..((........))...(((((..(--------(((.(((((((....((....))...)))))))..))))...(((((((((((((.....)).))))..)))))...)))))))... ( -31.10) >DroSim_CAF1 33110 112 - 1 CAGCAGCCCACAGCAACCACCCACU--------GCCCACCUGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGAUGUCA ..((((.................))--------))..(((((((....((....))...)))))))..(((((.((((....((((((.....)).))))((....)).)))).))))). ( -25.33) >DroEre_CAF1 13373 117 - 1 CGGCAACCCACA---GCCACCCACUGCCCACCUGCUCAUCUGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCA .(((........---)))((((..((((.(((((((...(((((.....))))).....)))))))..))))...(((....((((((.....)).))))((....)).))))))).... ( -35.80) >DroYak_CAF1 25410 111 - 1 ------CCAUCC---ACCACCCACUGCCCACCUGCUCAUCCGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCA ------......---..(((((..((((.(((((((.....(((........)))....)))))))..))))...(((....((((((.....)).))))((....)).))))))))... ( -31.70) >consensus CAGCAGCCCACAGC_ACCACCCACUG_CCACCUGCCCACCUGCUCAUCAGGUAGCUUUUAGCAGGUCCGGCAUACGGUAAAUUGUGCACAUUUUGACACAUCAUUUGACACCGGGUGUCA .................(((((...........(((.(((((((....((....))...)))))))..)))....(((((((((((((.....)).))))..)))))...)))))))... (-24.52 = -24.28 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:36 2006