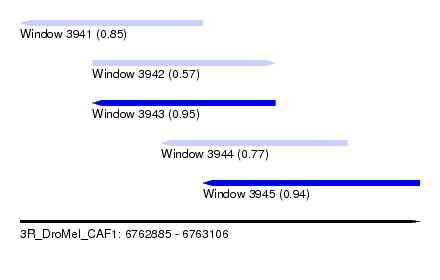
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,762,885 – 6,763,106 |
| Length | 221 |
| Max. P | 0.954844 |

| Location | 6,762,885 – 6,762,986 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762885 101 - 27905053 GU-----------------AAGUCCUUCAUUAAAUUGCAUUGGUGGCACCCAAAAAAUUUGGUUAGAUCCGAGCCACAAAUUGUGAAUCUGGUCACACAAAUAAGUCCUGAUGCAAGG ..-----------------...............((((((..(.(((..((((.....))))...((((.((..(((.....)))..)).))))..........))))..)))))).. ( -25.50) >DroSec_CAF1 27148 117 - 1 GCGGGAUGUCCCACAAAACGAGACCUUCAUUAAAUUGCAUUGGUGGCACGCAAAAAAUUUGGUUAAAUCCGAGCCAGAAAGUGUGAGUCU-AUCACACAAAGAAACCCUGAUGCAAGG (.(((....))).)....................((((((..(.((...........(((((((.......)))))))..((((((....-.)))))).......)))..)))))).. ( -32.10) >DroSim_CAF1 31472 118 - 1 GAGAGAUGUGUCACAAAACAAGUCCUUCAUUAAAUUGCAUUGGUGGCACGAAAAAAAUUUGGUUAAAUCCGAGCCACAAAGUGUGAAUCUGGUCACACAAAGAAACCCUGAUGCAAGG (((.(((.(((......))).))).)))......(((((((.(((((...........((((......)))))))))...((((((......))))))...........))))))).. ( -29.20) >consensus GAG_GAUGU__CACAAAACAAGUCCUUCAUUAAAUUGCAUUGGUGGCACGCAAAAAAUUUGGUUAAAUCCGAGCCACAAAGUGUGAAUCUGGUCACACAAAGAAACCCUGAUGCAAGG ..................................((((((..(((((...........((((......))))))))....((((((......)))))).........)..)))))).. (-21.90 = -22.23 + 0.33)

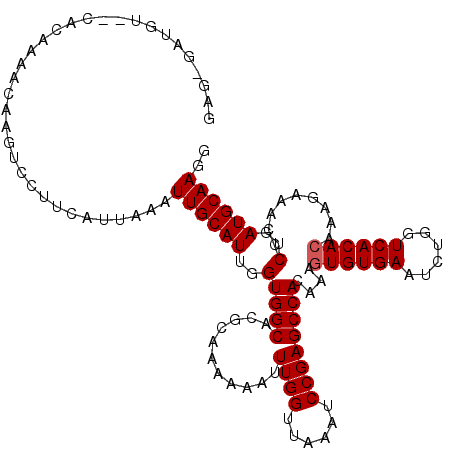
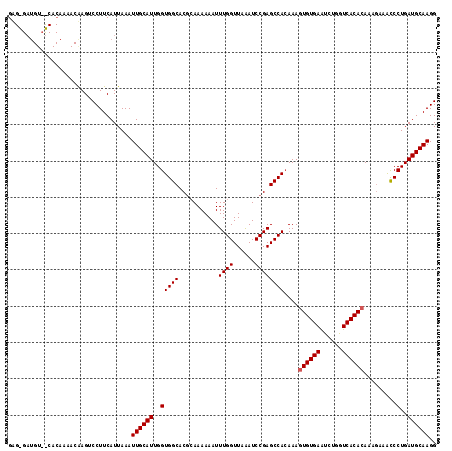
| Location | 6,762,925 – 6,763,026 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -22.55 |
| Energy contribution | -22.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762925 101 + 27905053 UGUGGCUCGGAUCUAACCAAAUUUUUUGGGUGCCACCAAUGCAAUUUAAUGAAGGACUU-----------------ACCAUUUAUUUCCCACAGUGCAUUGUGGCUGUGUGCCUUGAG ...(((.((.(.....((((.....))))..(((((.((((((....((((((((....-----------------.)).))))))........)))))))))))).)).)))..... ( -26.50) >DroSec_CAF1 27187 118 + 1 UCUGGCUCGGAUUUAACCAAAUUUUUUGCGUGCCACCAAUGCAAUUUAAUGAAGGUCUCGUUUUGUGGGACAUCCCGCCAUUUAUUUCCCACAGUGCAUUGUGGCUGUAUGCCUUAAG ...(((..((......))........((((.(((((.((((((....(((((.....)))))((((((((................))))))))))))))))))))))).)))..... ( -34.09) >DroSim_CAF1 31512 118 + 1 UGUGGCUCGGAUUUAACCAAAUUUUUUUCGUGCCACCAAUGCAAUUUAAUGAAGGACUUGUUUUGUGACACAUCUCUCCAUUUAUUUUCCACAGUGCAUUGUGGCUGUAUGCCUUAAG .(((((.((((...............)))).)))))(((((((....(((((((((..(((........)))....))).))))))........))))))).(((.....)))..... ( -28.36) >consensus UGUGGCUCGGAUUUAACCAAAUUUUUUGCGUGCCACCAAUGCAAUUUAAUGAAGGACUUGUUUUGUG__ACAUC_CACCAUUUAUUUCCCACAGUGCAUUGUGGCUGUAUGCCUUAAG ...(((..((......)).............(((((.((((((....((((((((......................)).))))))........))))))))))).....)))..... (-22.55 = -22.55 + 0.00)

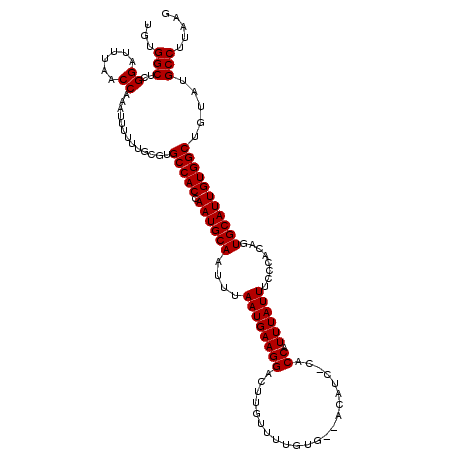
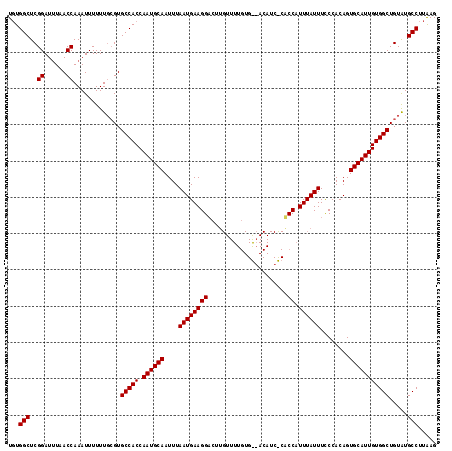
| Location | 6,762,925 – 6,763,026 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.35 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762925 101 - 27905053 CUCAAGGCACACAGCCACAAUGCACUGUGGGAAAUAAAUGGU-----------------AAGUCCUUCAUUAAAUUGCAUUGGUGGCACCCAAAAAAUUUGGUUAGAUCCGAGCCACA .....(((.....(((((((((((.((.((((..((.....)-----------------)..)))).))......)))))).)))))...........((((......)))))))... ( -26.50) >DroSec_CAF1 27187 118 - 1 CUUAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUGGCGGGAUGUCCCACAAAACGAGACCUUCAUUAAAUUGCAUUGGUGGCACGCAAAAAAUUUGGUUAAAUCCGAGCCAGA .....(((.....(((((((((((.(((((((.((..........)).)))))))....(((...))).......)))))).)))))...........((((......)))))))... ( -33.00) >DroSim_CAF1 31512 118 - 1 CUUAAGGCAUACAGCCACAAUGCACUGUGGAAAAUAAAUGGAGAGAUGUGUCACAAAACAAGUCCUUCAUUAAAUUGCAUUGGUGGCACGAAAAAAAUUUGGUUAAAUCCGAGCCACA .....(((.....(((((((((((.(((.....)))(((((((.(((.(((......))).))))))))))....)))))).)))))...........((((......)))))))... ( -33.70) >consensus CUUAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUGGAG_GAUGU__CACAAAACAAGUCCUUCAUUAAAUUGCAUUGGUGGCACGCAAAAAAUUUGGUUAAAUCCGAGCCACA .....(((.....(((((((((((..(((((..................................))))).....)))))).)))))...........((((......)))))))... (-23.57 = -23.35 + -0.22)

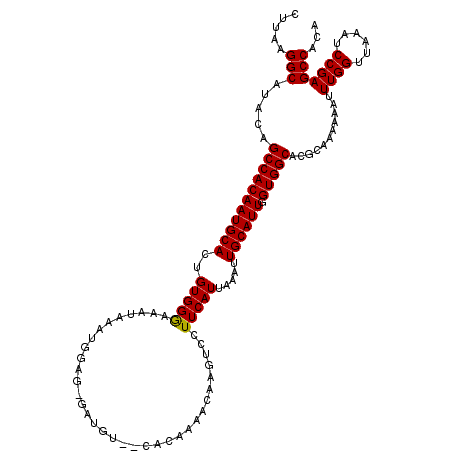
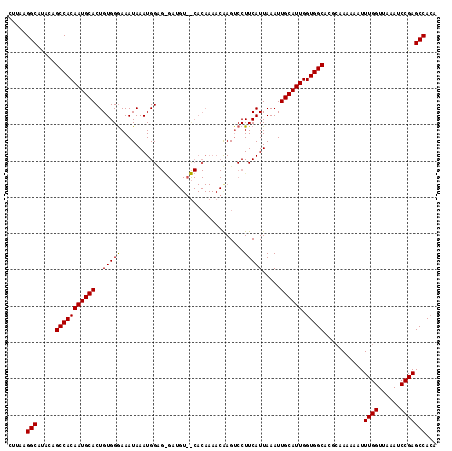
| Location | 6,762,963 – 6,763,066 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.24 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762963 103 - 27905053 CCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCACACAGCCACAAUGCACUGUGGGAAAUAAAUGGU-----------------AAGUCCUUCAUUAAAUUGCAU ..((((..(((((......))))).(((((...((((((......(((.....))).))))))...))))).......))))-----------------..................... ( -28.90) >DroSec_CAF1 27225 120 - 1 CCACCAGCCGCCACCCACUCGGUGACCCACUUAGCAUUGCCUUAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUGGCGGGAUGUCCCACAAAACGAGACCUUCAUUAAAUUGCAU ......((((.((((.....)))).(((((...((((((......(((.....))).))))))...))))).......))))(((....)))............................ ( -33.80) >DroSim_CAF1 31550 120 - 1 CCACCAGCCGCCGCCCACUGGUUGACCCACUUAGCAUUGCCUUAAGGCAUACAGCCACAAUGCACUGUGGAAAAUAAAUGGAGAGAUGUGUCACAAAACAAGUCCUUCAUUAAAUUGCAU ....((((((........))))))..((((...((((((......(((.....))).))))))...))))......(((((((.(((.(((......))).))))))))))......... ( -31.30) >DroEre_CAF1 11614 111 - 1 CCGUCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGUAUACAGUCGCAAUGCACUGUGGGAAACAAGUCGUGCGAUACAUUGCAAAAUGAUUCCUUCACA--------- .((.(...(((((......))))).(((((...(((((((..(..........)..)))))))...)))))......).))(((((....)))))...(((.....)))..--------- ( -30.40) >DroYak_CAF1 23735 111 - 1 CCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCAUACAGCCACAAUGCACUGUGCGAAAUAAGUGGUGGGAUGUGUUGCAAAAUUAUUCCUUCAUU--------- ......((.(((((((((..(....).(((((((((((((.....(((.....))).....)))..))))....))))))))))).)).)).)).................--------- ( -29.50) >consensus CCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUGGUGGGAUGUGUCACAAAACAAGUCCUUCAUUAAAUUGCAU .(((((..((((........)))).(((((...((((((......(((.....))).))))))...))))).......)))))..................................... (-20.40 = -21.24 + 0.84)

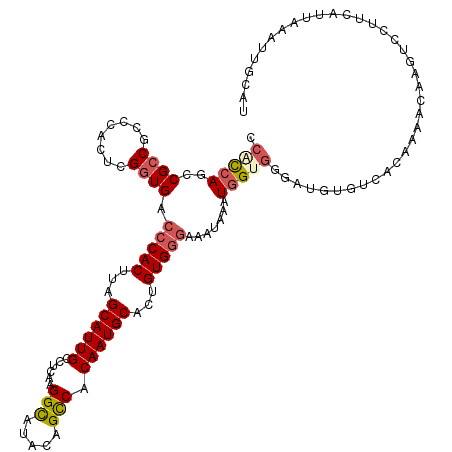
| Location | 6,762,986 – 6,763,106 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -25.09 |
| Energy contribution | -25.21 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6762986 120 - 27905053 ACCACUCAGCGGCCCCACUUCAUCAUUAAUGCACCGAAAUCCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCACACAGCCACAAUGCACUGUGGGAAAUAAAUG ........(((((.........((...........)).........)))))((((.....)))).(((((...((((((......(((.....))).))))))...)))))......... ( -29.57) >DroSec_CAF1 27265 120 - 1 ACCACUCAGCGGCCCCACUUCAUCAUUAAUGCACCGAAAUCCACCAGCCGCCACCCACUCGGUGACCCACUUAGCAUUGCCUUAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUG ........(((((.........((...........)).........)))))((((.....)))).(((((...((((((......(((.....))).))))))...)))))......... ( -29.97) >DroSim_CAF1 31590 120 - 1 ACCACUCAGCGGCCCCACUUCAUCAUUAAUGCACCGAAAUCCACCAGCCGCCGCCCACUGGUUGACCCACUUAGCAUUGCCUUAAGGCAUACAGCCACAAUGCACUGUGGAAAAUAAAUG ((((....(((((....................................)))))....))))....((((...((((((......(((.....))).))))))...)))).......... ( -24.92) >DroEre_CAF1 11645 120 - 1 ACCACUCAGCGGCCCCAUUUCAUCAUUAAUGCACCGAAAUCCGUCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGUAUACAGUCGCAAUGCACUGUGGGAAACAAGUC ........(((((..((((........))))....((......)).)))))((((.....)))).(((((...(((((((..(..........)..)))))))...)))))......... ( -32.40) >DroYak_CAF1 23766 120 - 1 ACCACUCAGCGGCCCCACUUCAUCAUUAAUGCACCGAAAUCCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCAUACAGCCACAAUGCACUGUGCGAAAUAAGUG ........(((((.........((...........)).........)))))((((.....))))...(((((((((((((.....(((.....))).....)))..))))....)))))) ( -24.37) >consensus ACCACUCAGCGGCCCCACUUCAUCAUUAAUGCACCGAAAUCCACCAGCCGCCGCCCACUCGGUGACCCACUUAGCAUUGCCUCAAGGCAUACAGCCACAAUGCACUGUGGGAAAUAAAUG ........(((((.........((...........)).........)))))((((.....)))).(((((...((((((......(((.....))).))))))...)))))......... (-25.09 = -25.21 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:31 2006